Circulating NK cells establish tissue residency upon acute infection of skin and mediate accelerated effector responses to secondary infection
- PMID: 38157853
- PMCID: PMC10783803
- DOI: 10.1016/j.immuni.2023.11.018
Circulating NK cells establish tissue residency upon acute infection of skin and mediate accelerated effector responses to secondary infection
Abstract
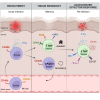
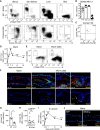
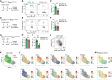
Natural killer (NK) cells are present in the circulation and can also be found residing in tissues, and these populations exhibit distinct developmental requirements and are thought to differ in terms of ontogeny. Here, we investigate whether circulating conventional NK (cNK) cells can develop into long-lived tissue-resident NK (trNK) cells following acute infections. We found that viral and bacterial infections of the skin triggered the recruitment of cNK cells and their differentiation into Tcf1hiCD69hi trNK cells that share transcriptional similarity with CD56brightTCF1hi NK cells in human tissues. Skin trNK cells arose from interferon (IFN)-γ-producing effector cells and required restricted expression of the transcriptional regulator Blimp1 to optimize Tcf1-dependent trNK cell formation. Upon secondary infection, trNK cells rapidly gained effector function and mediated an accelerated NK cell response. Thus, cNK cells redistribute and permanently position at sites of previous infection via a mechanism promoting tissue residency that is distinct from Hobit-dependent developmental paths of NK cells and ILC1 seeding tissues during ontogeny.
Keywords: Staphylococcus aureus; infection; innate immune memory; innate lymphoid cells; natural killer cells; tissue immunity; tissue-resident lymphocytes; trained immunity; vaccination; vaccinia virus.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





Comment in
-
On blood and tissue-resident natural killer cells.Immunity. 2024 Jan 9;57(1):6-8. doi: 10.1016/j.immuni.2023.12.013. Immunity. 2024. PMID: 38198854
References
-
- Mujal A.M., Delconte R.B., Sun J.C. Natural killer cells: from innate to adaptive features. Annu. Rev. Immunol. 2021;39:417–447. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

