Sterile protection against P. vivax malaria by repeated blood stage infection in the Aotus monkey model
- PMID: 38158220
- PMCID: PMC10756917
- DOI: 10.26508/lsa.202302524
Sterile protection against P. vivax malaria by repeated blood stage infection in the Aotus monkey model
Abstract
The malaria parasite Plasmodium vivax remains a major global public health challenge, and no vaccine is approved for use in humans. Here, we assessed whether P. vivax strain-transcendent immunity can be achieved by repeated infection in Aotus monkeys. Sterile immunity was achieved after two homologous infections, whereas subsequent heterologous challenge provided only partial protection. IgG levels based on P. vivax lysate ELISA and protein microarray increased with repeated infections and correlated with the level of homologous protection. Parasite transcriptional profiles provided no evidence of major antigenic switching upon homologous or heterologous challenge. However, we observed significant sequence diversity and transcriptional differences in the P. vivax core gene repertoire between the two strains used in the study, suggesting that partial protection upon heterologous challenge is due to molecular differences between strains rather than immune evasion by antigenic switching. Our study demonstrates that sterile immunity against P. vivax can be achieved by repeated homologous blood stage infection in Aotus monkeys, thus providing a benchmark to test the efficacy of candidate blood stage P. vivax malaria vaccines.
© 2023 Obaldía et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

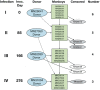

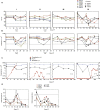

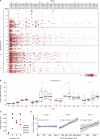

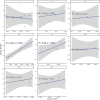



Similar articles
-
A Plasmodium vivax Plasmid DNA- and Adenovirus-Vectored Malaria Vaccine Encoding Blood-Stage Antigens AMA1 and MSP142 in a Prime/Boost Heterologous Immunization Regimen Partially Protects Aotus Monkeys against Blood-Stage Challenge.Clin Vaccine Immunol. 2017 Apr 5;24(4):e00539-16. doi: 10.1128/CVI.00539-16. Print 2017 Apr. Clin Vaccine Immunol. 2017. PMID: 28179404 Free PMC article.
-
Protective efficacy of a Plasmodium vivax circumsporozoite protein-based vaccine in Aotus nancymaae is associated with antibodies to the repeat region.PLoS Negl Trop Dis. 2014 Oct 16;8(10):e3268. doi: 10.1371/journal.pntd.0003268. eCollection 2014 Oct. PLoS Negl Trop Dis. 2014. PMID: 25329054 Free PMC article.
-
Immune responses and protection of Aotus monkeys immunized with irradiated Plasmodium vivax sporozoites.Am J Trop Med Hyg. 2011 Feb;84(2 Suppl):43-50. doi: 10.4269/ajtmh.2011.09-0759. Am J Trop Med Hyg. 2011. PMID: 21292877 Free PMC article.
-
Plasmodium vivax malaria vaccine development.Mol Immunol. 2001 Dec;38(6):443-55. doi: 10.1016/s0161-5890(01)00080-3. Mol Immunol. 2001. PMID: 11741694 Review.
-
Plasmodium vivax vaccine: What is the best way to go?Front Immunol. 2023 Jan 16;13:910236. doi: 10.3389/fimmu.2022.910236. eCollection 2022. Front Immunol. 2023. PMID: 36726991 Free PMC article. Review.
Cited by
-
The human malaria-Aotus monkey model: a historical perspective in antimalarial chemotherapy research at the Gorgas Memorial Laboratory-Panama.Antimicrob Agents Chemother. 2024 Jul 9;68(7):e0033824. doi: 10.1128/aac.00338-24. Epub 2024 Jun 5. Antimicrob Agents Chemother. 2024. PMID: 38837364 Free PMC article. Review.
References
-
- WHO Team (2021) World Malaria Report 2021. Geneva, Switzerland: WHO.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
