Quantifying the Impact of the Peptide Identification Framework on the Results of Fast Photochemical Oxidation of Protein Analysis
- PMID: 38158558
- PMCID: PMC10845142
- DOI: 10.1021/acs.jproteome.3c00390
Quantifying the Impact of the Peptide Identification Framework on the Results of Fast Photochemical Oxidation of Protein Analysis
Abstract
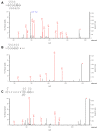
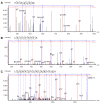
Fast Photochemical Oxidation of Proteins (FPOP) is a promising technique for studying protein structure and dynamics. The quality of insight provided by FPOP depends on the reliability of the determination of the modification site. This study investigates the performance of two search engines, Mascot and PEAKS, for the data processing of FPOP analyses. Comparison of Mascot and PEAKS of the hemoglobin--haptoglobin Bruker timsTOF data set (PXD021621) revealed greater consistency in the Mascot identification of modified peptides, with around 26% of the IDs being mutual for all three replicates, compared to approximately 22% for PEAKS. The intersection between Mascot and PEAKS results revealed a limited number (31%) of shared modified peptides. Principal Component Analysis (PCA) using the peptide-spectrum match (PSM) score, site probability, and peptide intensity was applied to evaluate the results, and the analyses revealed distinct clusters of modified peptides. Mascot showed the ability to assess confident site determination, even with lower PSM scores. However, high PSM scores from PEAKS did not guarantee a reliable determination of the modification site. Fragmentation coverage of the modification position played a crucial role in Mascot assignments, while the AScore localizations from PEAKS often become ambiguous because the software employs MS/MS merging.
Keywords: FPOP; search engine; structural proteomics.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


Similar articles
-
Enhanced peptide identification by electron transfer dissociation using an improved Mascot Percolator.Mol Cell Proteomics. 2012 Aug;11(8):478-91. doi: 10.1074/mcp.O111.014522. Epub 2012 Apr 6. Mol Cell Proteomics. 2012. PMID: 22493177 Free PMC article.
-
In-depth analysis of protein inference algorithms using multiple search engines and well-defined metrics.J Proteomics. 2017 Jan 6;150:170-182. doi: 10.1016/j.jprot.2016.08.002. Epub 2016 Aug 4. J Proteomics. 2017. PMID: 27498275
-
Altered Mascot search results by changing the m/z range of MS/MS spectra: analysis and potential applications.Anal Bioanal Chem. 2011 Jun;400(8):2339-47. doi: 10.1007/s00216-010-4572-0. Epub 2010 Dec 15. Anal Bioanal Chem. 2011. PMID: 21161510
-
Fast Photochemical Oxidation of Proteins Coupled with Mass Spectrometry.Protein Pept Lett. 2019;26(1):27-34. doi: 10.2174/0929866526666181128124554. Protein Pept Lett. 2019. PMID: 30484399 Free PMC article. Review.
-
Mass Spectrometry-Based Fast Photochemical Oxidation of Proteins (FPOP) for Higher Order Structure Characterization.Acc Chem Res. 2018 Mar 20;51(3):736-744. doi: 10.1021/acs.accounts.7b00593. Epub 2018 Feb 16. Acc Chem Res. 2018. PMID: 29450991 Free PMC article. Review.
Cited by
-
Data-Independent Acquisition Represents a Promising Alternative for Fast Photochemical Oxidation of Proteins (FPOP) Samples Analysis.Anal Chem. 2024 Jul 16;96(28):11273-11279. doi: 10.1021/acs.analchem.4c01084. Epub 2024 Jul 5. Anal Chem. 2024. PMID: 38967040 Free PMC article.
-
Covalent Labeling Automated Data Analysis Platform for High Throughput in R (coADAPTr): A Proteome-Wide Data Analysis Platform for Covalent Labeling Experiments.J Am Soc Mass Spectrom. 2024 Dec 4;35(12):3301-3307. doi: 10.1021/jasms.4c00196. Epub 2024 Oct 2. J Am Soc Mass Spectrom. 2024. PMID: 39356573 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

