Phosphoproteomic Characterization and Kinase Signature Predict Response to Venetoclax Plus 3+7 Chemotherapy in Acute Myeloid Leukemia
- PMID: 38161214
- PMCID: PMC10953567
- DOI: 10.1002/advs.202305885
Phosphoproteomic Characterization and Kinase Signature Predict Response to Venetoclax Plus 3+7 Chemotherapy in Acute Myeloid Leukemia
Abstract
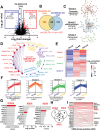
Resistance to chemotherapy remains a formidable obstacle in acute myeloid leukemia (AML) therapeutic management, necessitating the exploration of optimal strategies to maximize therapeutic benefits. Venetoclax with 3+7 daunorubicin and cytarabine (DAV regimen) in young adult de novo AML patients is evaluated. 90% of treated patients achieved complete remission, underscoring the potential of this regimen as a compelling therapeutic intervention. To elucidate underlying mechanisms governing response to DAV in AML, quantitative phosphoproteomics to discern distinct molecular signatures characterizing a subset of DAV-sensitive patients is used. Cluster analysis reveals an enrichment of phosphoproteins implicated in chromatin organization and RNA processing within DAV-susceptible and DA-resistant AML patients. Furthermore, kinase activity profiling identifies AURKB as a candidate indicator of DAV regimen efficacy in DA-resistant AML due to AURKB activation. Intriguingly, AML cells overexpressing AURKB exhibit attenuated MCL-1 expression, rendering them receptive to DAV treatment and maintaining them resistant to DA treatment. Moreover, the dataset delineates a shared kinase, AKT1, associated with DAV response. Notably, AKT1 inhibition augments the antileukemic efficacy of DAV treatment in AML. Overall, this phosphoproteomic study identifies the role of AURKB as a predictive biomarker for DA, but not DAV, resistance and proposes a promising strategy to counteract therapy resistance in AML.
Keywords: AKT1; AURKB; acute myeloid leukemia; drug resistance; venetoclax.
© 2023 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
MeSH terms
Substances
Grants and funding
- 226-2022-00003/Fundamental Research Funds for the Central Universities
- 22120230064/Fundamental Research Funds for the Central Universities
- 23141902700/2023 Science and Technology Innovation Plan of Shanghai Science and Technology Commission
- 82370162/National Natural Science Foundation of China
- 81820108004/National Natural Science Foundation of China
- 82170144/National Natural Science Foundation of China
- JNL-2022034C/Research Project of Jinan Microecological Biomedicine Shandong Laboratory
- 2022C03005/Key Research and Development Program of Zhejiang Province
- 2021C03123/Key Research and Development Program of Zhejiang Province
- Y23H080018/Natural Science Foundation of Zhejiang Province
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
