Reference genome of the nutrition-rich orphan crop chia (Salvia hispanica) and its implications for future breeding
- PMID: 38162307
- PMCID: PMC10757625
- DOI: 10.3389/fpls.2023.1272966
Reference genome of the nutrition-rich orphan crop chia (Salvia hispanica) and its implications for future breeding
Abstract
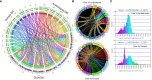
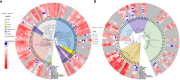
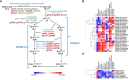
Chia (Salvia hispanica L.) is one of the most popular nutrition-rich foods and pseudocereal crops of the family Lamiaceae. Chia seeds are a rich source of proteins, polyunsaturated fatty acids (PUFAs), dietary fibers, and antioxidants. In this study, we present the assembly of the chia reference genome, which spans 303.6 Mb and encodes 48,090 annotated protein-coding genes. Our analysis revealed that ~42% of the chia genome harbors repetitive content, and identified ~3 million single nucleotide polymorphisms (SNPs) and 15,380 simple sequence repeat (SSR) marker sites. By investigating the chia transcriptome, we discovered that ~44% of the genes undergo alternative splicing with a higher frequency of intron retention events. Additionally, we identified chia genes associated with important nutrient content and quality traits, such as the biosynthesis of PUFAs and seed mucilage fiber (dietary fiber) polysaccharides. Notably, this is the first report of in-silico annotation of a plant genome for protein-derived small bioactive peptides (biopeptides) associated with improving human health. To facilitate further research and translational applications of this valuable orphan crop, we have developed the Salvia genomics database (SalviaGDB), accessible at https://salviagdb.org.
Keywords: Salvia hispanica; biopeptide; chia; lectins; polyunsaturated fatty acid (PUFA); reference genome; seed mucilage; terpene synthase.
Copyright © 2023 Gupta, Geniza, Elser, Al-Bader, Baschieri, Phillips, Haq, Preece, Naithani and Jaiswal.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures



Similar articles
-
Chia (Salvia hispanica) Gene Expression Atlas Elucidates Dynamic Spatio-Temporal Changes Associated With Plant Growth and Development.Front Plant Sci. 2021 Jul 20;12:667678. doi: 10.3389/fpls.2021.667678. eCollection 2021. Front Plant Sci. 2021. PMID: 34354718 Free PMC article.
-
Multi-omic analyses reveal the unique properties of chia (Salvia hispanica) seed metabolism.Commun Biol. 2023 Aug 7;6(1):820. doi: 10.1038/s42003-023-05192-4. Commun Biol. 2023. PMID: 37550387 Free PMC article.
-
Exploring triacylglycerol biosynthetic pathway in developing seeds of Chia (Salvia hispanica L.): a transcriptomic approach.PLoS One. 2015 Apr 13;10(4):e0123580. doi: 10.1371/journal.pone.0123580. eCollection 2015. PLoS One. 2015. PMID: 25875809 Free PMC article.
-
An overview of chia seed (Salvia hispanica L.) bioactive peptides' derivation and utilization as an emerging nutraceutical food.Front Biosci (Landmark Ed). 2021 Sep 30;26(9):643-654. doi: 10.52586/4973. Front Biosci (Landmark Ed). 2021. PMID: 34590473 Review.
-
Chia (Salvia hispanica L.) Seed Germination: a Brief Review.Plant Foods Hum Nutr. 2022 Dec;77(4):485-494. doi: 10.1007/s11130-022-01011-z. Epub 2022 Sep 9. Plant Foods Hum Nutr. 2022. PMID: 36083408 Review.
Cited by
-
Transcriptional Modulation during Photomorphogenesis in Rice Seedlings.Genes (Basel). 2024 Aug 14;15(8):1072. doi: 10.3390/genes15081072. Genes (Basel). 2024. PMID: 39202430 Free PMC article.
-
Application of machine learning and genomics for orphan crop improvement.Nat Commun. 2025 Jan 24;16(1):982. doi: 10.1038/s41467-025-56330-x. Nat Commun. 2025. PMID: 39856113 Free PMC article. Review.
-
Haplotype-resolved chromosome-level genome sequence of Elsholtzia splendens (Nakai ex F.Maek.).Sci Data. 2025 May 20;12(1):827. doi: 10.1038/s41597-025-05214-2. Sci Data. 2025. PMID: 40394069 Free PMC article.
-
Chia (Salvia hispanica L.), a functional 'superfood': new insights into its botanical, genetic and nutraceutical characteristics.Ann Bot. 2024 Nov 13;134(5):725-746. doi: 10.1093/aob/mcae123. Ann Bot. 2024. PMID: 39082745 Free PMC article. Review.
-
From 'Farm to Fork': Exploring the Potential of Nutrient-Rich and Stress-Resilient Emergent Crops for Sustainable and Healthy Food in the Mediterranean Region in the Face of Climate Change Challenges.Plants (Basel). 2024 Jul 11;13(14):1914. doi: 10.3390/plants13141914. Plants (Basel). 2024. PMID: 39065441 Free PMC article. Review.
References
-
- ALBERTO C. M., SANSO A. M., XIFREDA C. C. (2003). Chromosomal studies in species of Salvia (Lamiaceae) from Argentina. Bot. J. Linn. Soc. 141, 483–490. doi: 10.1046/j.1095-8339.2003.t01-1-00178.x - DOI
-
- Alejo-Jacuinde G., Nájera-González H. R., Chávez Montes R. A., Gutierrez Reyes C. D., Barragán-Rosillo A. C., Perez Sanchez B., et al. . (2023). Multi-omic analyses reveal the unique properties of chia (Salvia hispanica) seed metabolism. Commun. Biol. 6, 1–13. doi: 10.1038/s42003-023-05192-4 - DOI - PMC - PubMed
-
- Alfredo V.-O., Gabriel R.-R., Luis C.-G., David B.-A. (2009). Physicochemical properties of a fibrous fraction from chia (Salvia hispanica L.). LWT Food Sci. Technol. 42, 168–173. doi: 10.1016/j.lwt.2008.05.012 - DOI
Associated data
LinkOut - more resources
Full Text Sources
Miscellaneous

