Deep learning-based breast region segmentation in raw and processed digital mammograms: generalization across views and vendors
- PMID: 38162417
- PMCID: PMC10753125
- DOI: 10.1117/1.JMI.11.1.014001
Deep learning-based breast region segmentation in raw and processed digital mammograms: generalization across views and vendors
Abstract
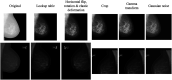
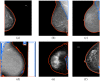
Purpose: We developed a segmentation method suited for both raw (for processing) and processed (for presentation) digital mammograms (DMs) that is designed to generalize across images acquired with systems from different vendors and across the two standard screening views.
Approach: A U-Net was trained to segment mammograms into background, breast, and pectoral muscle. Eight different datasets, including two previously published public sets and six sets of DMs from as many different vendors, were used, totaling 322 screen film mammograms (SFMs) and 4251 DMs (2821 raw/processed pairs and 1430 only processed) from 1077 different women. Three experiments were done: first training on all SFM and processed images, second also including all raw images in training, and finally testing vendor generalization by leaving one dataset out at a time.
Results: The model trained on SFM and processed mammograms achieved a good overall performance regardless of projection and vendor, with a mean (±std. dev.) dice score of for all datasets combined. When raw images were included in training, the mean (±std. dev.) dice score for the raw images was and for the processed images was . Testing on a dataset with processed DMs from a vendor that was excluded from training resulted in a difference in mean dice varying between to from that of the fully trained model.
Conclusions: The proposed segmentation method yields accurate overall segmentation results for both raw and processed mammograms independent of view and vendor. The code and model weights are made available.
Keywords: U-Net; deep convolutional neural network; mammography; pectoral muscle; segmentation.
© 2023 The Authors.
Figures

References
-
- Zebari D. A., et al. , “Improved threshold based and trainable fully automated segmentation for breast cancer boundary and pectoral muscle in mammogram images,” IEEE Access 8, 203097–203116 (2020).10.1109/ACCESS.2020.3036072 - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

