Discovering common pathogenetic processes between COVID-19 and tuberculosis by bioinformatics and system biology approach
- PMID: 38162574
- PMCID: PMC10757339
- DOI: 10.3389/fcimb.2023.1280223
Discovering common pathogenetic processes between COVID-19 and tuberculosis by bioinformatics and system biology approach
Abstract
Introduction: The coronavirus disease 2019 (COVID-19) pandemic, stemming from the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has persistently threatened the global health system. Meanwhile, tuberculosis (TB) caused by Mycobacterium tuberculosis (M. tuberculosis) still continues to be endemic in various regions of the world. There is a certain degree of similarity between the clinical features of COVID-19 and TB, but the underlying common pathogenetic processes between COVID-19 and TB are not well understood.
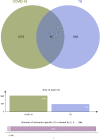
Methods: To elucidate the common pathogenetic processes between COVID-19 and TB, we implemented bioinformatics and systematic research to obtain shared pathways and molecular biomarkers. Here, the RNA-seq datasets (GSE196822 and GSE126614) are used to extract shared differentially expressed genes (DEGs) of COVID-19 and TB. The common DEGs were used to identify common pathways, hub genes, transcriptional regulatory networks, and potential drugs.
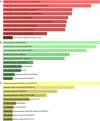
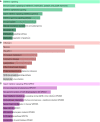
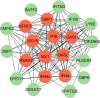
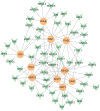
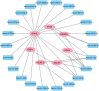
Results: A total of 96 common DEGs were selected for subsequent analyses. Functional enrichment analyses showed that viral genome replication and immune-related pathways collectively contributed to the development and progression of TB and COVID-19. Based on the protein-protein interaction (PPI) network analysis, we identified 10 hub genes, including IFI44L, ISG15, MX1, IFI44, OASL, RSAD2, GBP1, OAS1, IFI6, and HERC5. Subsequently, the transcription factor (TF)-gene interaction and microRNA (miRNA)-gene coregulatory network identified 61 TFs and 29 miRNAs. Notably, we identified 10 potential drugs to treat TB and COVID-19, namely suloctidil, prenylamine, acetohexamide, terfenadine, prochlorperazine, 3'-azido-3'-deoxythymidine, chlorophyllin, etoposide, clioquinol, and propofol.
Conclusion: This research provides novel strategies and valuable references for the treatment of tuberculosis and COVID-19.
Keywords: SARS-CoV-2; differentially expressed genes; drug molecule; hub gene; protein-protein interaction (PPI); tuberculosis.
Copyright © 2023 Huang, He, Zhou, Pan, He, Du, Yu, Jiang, Li, Yuan and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
IFI44 is an immune evasion biomarker for SARS-CoV-2 and Staphylococcus aureus infection in patients with RA.Front Immunol. 2022 Sep 15;13:1013322. doi: 10.3389/fimmu.2022.1013322. eCollection 2022. Front Immunol. 2022. PMID: 36189314 Free PMC article.
-
Bioinformatics and system biology approach to discover the common pathogenetic processes between COVID-19 and chronic hepatitis B.PLoS One. 2025 May 23;20(5):e0323708. doi: 10.1371/journal.pone.0323708. eCollection 2025. PLoS One. 2025. PMID: 40408617 Free PMC article.
-
Investigation of the relationship between COVID-19 and pancreatic cancer using bioinformatics and systems biology approaches.Medicine (Baltimore). 2024 Aug 2;103(31):e39057. doi: 10.1097/MD.0000000000039057. Medicine (Baltimore). 2024. PMID: 39093763 Free PMC article.
-
Comprehensive analyses of potential key genes in active tuberculosis: A systematic review.Medicine (Baltimore). 2021 Jul 30;100(30):e26582. doi: 10.1097/MD.0000000000026582. Medicine (Baltimore). 2021. PMID: 34397688 Free PMC article.
-
Gene expression profiling of host lipid metabolism in SARS-CoV-2 infected patients: a systematic review and integrated bioinformatics analysis.BMC Infect Dis. 2024 Jan 23;24(1):124. doi: 10.1186/s12879-024-08983-0. BMC Infect Dis. 2024. PMID: 38263024 Free PMC article.
Cited by
-
Exploration of the link between COVID-19 and gastric cancer from the perspective of bioinformatics and systems biology.Front Med (Lausanne). 2024 Sep 20;11:1428973. doi: 10.3389/fmed.2024.1428973. eCollection 2024. Front Med (Lausanne). 2024. PMID: 39371335 Free PMC article.
-
Quantitative Proteomic Analysis of Macrophages Infected with Trypanosoma cruzi Reveals Different Responses Dependent on the SLAMF1 Receptor and the Parasite Strain.Int J Mol Sci. 2024 Jul 8;25(13):7493. doi: 10.3390/ijms25137493. Int J Mol Sci. 2024. PMID: 39000601 Free PMC article.
-
Viperin: A Multifunctional Protein in Antiviral Immunity and Disease Pathogenesis.Pathogens. 2025 May 21;14(5):510. doi: 10.3390/pathogens14050510. Pathogens. 2025. PMID: 40430829 Free PMC article. Review.
-
Prediction of tuberculosis treatment outcomes using biochemical makers with machine learning.BMC Infect Dis. 2025 Feb 17;25(1):229. doi: 10.1186/s12879-025-10609-y. BMC Infect Dis. 2025. PMID: 39962412 Free PMC article.
-
Discovering common pathogenetic processes between periodontitis and Alzheimer's disease by bioinformatics and system biology approach.BMC Oral Health. 2024 Sep 12;24(1):1074. doi: 10.1186/s12903-024-04775-9. BMC Oral Health. 2024. PMID: 39266981 Free PMC article.
References
-
- Al-Mustanjid M., Mahmud S. M. H., Royel M. R. I., Rahman M. H., Islam T., Rahman M. R., et al. . (2020). Detection of molecular signatures and pathways shared in inflammatory bowel disease and colorectal cancer: A bioinformatics and systems biology approach. Genomics 112 (5), 3416–3426. doi: 10.1016/j.ygeno.2020.06.001 - DOI - PubMed
-
- Banday A. R., Stanifer M. L., Florez-Vargas O., Onabajo O. O., Papenberg B. W., Zahoor M. A., et al. . (2022). Genetic regulation of OAS1 nonsense-mediated decay underlies association with COVID-19 hospitalization in patients of European and African ancestries. Nat. Genet. 54 (8), 1103–1116. doi: 10.1038/s41588-022-01113-z - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

