Leveraging a gene signature associated with disulfidptosis identified by machine learning to forecast clinical outcomes, immunological heterogeneities, and potential therapeutic targets within lower-grade glioma
- PMID: 38162649
- PMCID: PMC10757341
- DOI: 10.3389/fimmu.2023.1294459
Leveraging a gene signature associated with disulfidptosis identified by machine learning to forecast clinical outcomes, immunological heterogeneities, and potential therapeutic targets within lower-grade glioma
Abstract
Background: Disulfidptosis, a newly defined type of programmed cell death, has emerged as a significant regulatory process in the development and advancement of malignant tumors, such as lower-grade glioma (LGG). Nevertheless, the precise biological mechanisms behind disulfidptosis in LGG are yet to be revealed, considering the limited research conducted in this field.
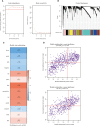
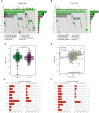
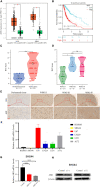
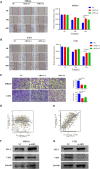
Methods: We obtained LGG data from the TCGA and CGGA databases and performed comprehensive weighted co-expression network analysis, single-sample gene set enrichment analysis, and transcriptome differential expression analyses. We discovered nine genes associated with disulfidptosis by employing machine learning methods like Cox regression, LASSO regression, and SVM-RFE. These were later used to build a predictive model for patients with LGG. To confirm the expression level, functional role, and impact on disulfidptosis of ABI3, the pivotal gene of the model, validation experiments were carried out in vitro.
Results: The developed prognostic model successfully categorized LGG patients into two distinct risk groups: high and low. There was a noticeable difference in the time the groups survived, which was statistically significant. The model's predictive accuracy was substantiated through two independent external validation cohorts. Additional evaluations of the immune microenvironment and the potential for immunotherapy indicated that this risk classification could function as a practical roadmap for LGG treatment using immune-based therapies. Cellular experiments demonstrated that suppressing the crucial ABI3 gene in the predictive model significantly reduced the migratory and invasive abilities of both SHG44 and U251 cell lines while also triggering cytoskeletal retraction and increased cell pseudopodia.
Conclusion: The research suggests that the prognostic pattern relying on genes linked to disulfidptosis can provide valuable insights into the clinical outcomes, tumor characteristics, and immune alterations in patients with LGG. This could pave the way for early interventions and suggests that ABI3 might be a potential therapeutic target for disulfidptosis.
Keywords: ABI3; disulfidptosis; lower-grade glioma; prognostic signature; tumor microenvironment.
Copyright © 2023 Zhou, Cao, Liu, Wang, Kuang, Zhou, Chen, Cheng, Huang, Zhang, Jiang, Wang and Ren.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures










Similar articles
-
A disulfidptosis-associated long noncoding RNA signature to predict low-grade glioma classification, prognosis, tumor microenvironment, and therapy regimens: Observational study.Medicine (Baltimore). 2024 Aug 23;103(34):e39316. doi: 10.1097/MD.0000000000039316. Medicine (Baltimore). 2024. PMID: 39183405 Free PMC article.
-
Recurrence- and Malignant Progression-Associated Biomarkers in Low-Grade Gliomas and Their Roles in Immunotherapy.Front Immunol. 2022 May 23;13:899710. doi: 10.3389/fimmu.2022.899710. eCollection 2022. Front Immunol. 2022. PMID: 35677036 Free PMC article.
-
Multiomics Analysis of Disulfidptosis Patterns and Integrated Machine Learning to Predict Immunotherapy Response in Lung Adenocarcinoma.Curr Med Chem. 2024;31(25):4034-4055. doi: 10.2174/0109298673313281240425050032. Curr Med Chem. 2024. PMID: 38685772
-
Identification of a novel cuproptosis-related gene signature and integrative analyses in patients with lower-grade gliomas.Front Immunol. 2022 Aug 15;13:933973. doi: 10.3389/fimmu.2022.933973. eCollection 2022. Front Immunol. 2022. PMID: 36045691 Free PMC article. Review.
-
Development and experimental verification of a prognosis model for disulfidptosis-associated genes in HNSCC.Medicine (Baltimore). 2024 Mar 22;103(12):e37308. doi: 10.1097/MD.0000000000037308. Medicine (Baltimore). 2024. PMID: 38518012 Free PMC article. Review.
Cited by
-
Machine learning-derived prognostic signature integrating programmed cell death and mitochondrial function in renal clear cell carcinoma: identification of PIF1 as a novel target.Cancer Immunol Immunother. 2025 Feb 25;74(4):113. doi: 10.1007/s00262-025-03967-8. Cancer Immunol Immunother. 2025. PMID: 39998680 Free PMC article.
-
Prognostic significance and multivariate modeling of COL4A family genes and HMGA2 in glioma.Front Pharmacol. 2025 Apr 25;16:1591932. doi: 10.3389/fphar.2025.1591932. eCollection 2025. Front Pharmacol. 2025. PMID: 40351420 Free PMC article.
-
Interrogation of macrophage-related prognostic signatures reveals a potential immune-mediated therapy strategy by histone deacetylase inhibition in glioma.Front Oncol. 2025 Jun 6;15:1554845. doi: 10.3389/fonc.2025.1554845. eCollection 2025. Front Oncol. 2025. PMID: 40548122 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

