Lineage tracing of T cell differentiation from T-iPSC by 2D feeder-free culture and 3D organoid culture
- PMID: 38162650
- PMCID: PMC10757342
- DOI: 10.3389/fimmu.2023.1303713
Lineage tracing of T cell differentiation from T-iPSC by 2D feeder-free culture and 3D organoid culture
Abstract
Introduction: T cells induced from induced pluripotent stem cells(iPSCs) derived from antigen-specific T cells (T-iPS-T cells) are an attractive tool for T cell immunotherapy. The induction of cytotoxic T-iPS-T cells is well established in feeder-free condition for the aim of off-the-shelf production, however, the induction of helper T-iPS-T cells remains challenging.
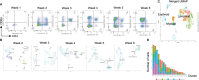
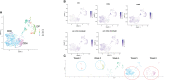
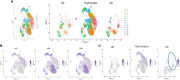
Methods: We analyzed T-iPS-T cells matured in 3D organoid culture at different steps in the culture process at the single-cell level. T-iPS-T cell datasets were merged with an available human thymocyte dataset based in single-cell RNA sequencing (scRNA-seq). Particularly, we searched for genes crucial for generation CD4+ T-iPS-T cells by comparing T-iPS-T cells established in 2D feeder-free or 3D organoid culture.
Results: The scRNA-seq data indicated that T-iPS-T cells are similar to T cells transitioning to human thymocytes, with SELENOW, GIMAP4, 7, SATB1, SALMF1, IL7R, SYTL2, S100A11, STAT1, IFITM1, LZTFL1 and SOX4 identified as candidate genes for the 2D feeder-free induction of CD4+ T-iPS-T cells.
Discussion: This study provides single cell transcriptome datasets of iPS-T cells and leads to further analysis for CD4+ T cell generation from T-iPSCs.
Keywords: 3D organoid; CD4; T cell differentiation; iPSC; scRNA seq.
Copyright © 2023 Ishiguro, Iriguchi, Asano, Shinohara, Shiina, Arima, Kassai, Sakai, Obama and Kaneko.
Conflict of interest statement
YK, SuA and TS are employees of Takeda Pharmaceutical Co. Ltd. ShA is an employee of Axcelead Drug Discovery Partners, Inc. SK is a founder, shareholder, and director at Thyas Co., Ltd. and received research fundings from Takeda Pharmaceutical Co., Ltd., Astellas Co., Ltd., Terumo Co., Ltd., Mitsui-soko Co., Ltd., Kotai Bio Co., Ltd.,and Thyas Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

