Urinary PART1 and PLA2R1 Could Potentially Serve as Diagnostic Markers for Diabetic Kidney Disease Patients
- PMID: 38162802
- PMCID: PMC10757812
- DOI: 10.2147/DMSO.S445341
Urinary PART1 and PLA2R1 Could Potentially Serve as Diagnostic Markers for Diabetic Kidney Disease Patients
Abstract
Background: Diabetic kidney disease (DKD) is a chronic renal disease which could eventually develop into renal failure. Though albuminuria and estimated glomerular filtration rate (eGFR) are helpful for the diagnosis of DKD, the lack of specific biomarkers reduces the efficiency of therapeutic interventions.
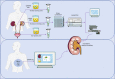
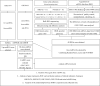
Methods: Based on bulk-seq of 56 urine samples collected at different time points (including 11 acquired from DKD patients and 11 from healthy controls), in corporation of scRNA-seq data of urine samples and snRNA-seq data of renal punctures from DKD patients (retrieved from NCBI GEO Omnibus), urine-kidney specific genes were identified by Multiple Biological Information methods.
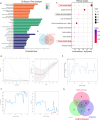
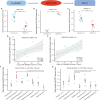
Results: Forty urine-kidney specific genes/differentially expressed genes (DEGs) were identified to be highly related to kidney injury and proteinuria for the DKD patients. Most of these genes participate in regulating glucagon and apoptosis, among which, urinary PART1 (mainly derived from distal tubular cells) and PLA2R1 (podocyte cell surface marker) could be used together for the early diagnosis of DKD. Moreover, urinary PART1 was significantly associated with multiple clinical indicators, and remained stable over time in urine.
Conclusion: Urinary PART1 and PLA2R1 could be shed lights on the discovery and development of non-invasive diagnostic method for DKD, especially in early stages.
Keywords: biomarker; diabetic kidney disease; diagnosis; non-invasive; urine.
© 2023 Ye et al.
Conflict of interest statement
The authors report no conflicts of interest in this work.
Figures




Similar articles
-
Association of urinary acidification function with the progression of diabetic kidney disease in patients with type 2 diabetes.J Diabetes Complications. 2019 Nov;33(11):107419. doi: 10.1016/j.jdiacomp.2019.107419. Epub 2019 Aug 19. J Diabetes Complications. 2019. PMID: 31473080
-
Diagnostic value of quantitative contrast-enhanced ultrasound (CEUS) for early detection of renal hyperperfusion in diabetic kidney disease.J Nephrol. 2015 Dec;28(6):669-78. doi: 10.1007/s40620-015-0183-3. Epub 2015 Feb 25. J Nephrol. 2015. PMID: 25712236
-
Clinical significance of UbA52 level in the urine of patients with type 2 diabetes mellitus and diabetic kidney disease.Nefrologia (Engl Ed). 2021 Sep-Oct;41(5):548-555. doi: 10.1016/j.nefroe.2021.11.009. Nefrologia (Engl Ed). 2021. PMID: 36165137
-
The Role of Podocytes and Podocyte-Associated Biomarkers in Diagnosis and Treatment of Diabetic Kidney Disease.J Endocr Soc. 2020 Mar 5;4(4):bvaa029. doi: 10.1210/jendso/bvaa029. eCollection 2020 Apr 1. J Endocr Soc. 2020. PMID: 32232184 Free PMC article. Review.
-
Novel biomarkers for prognosticating diabetic kidney disease progression.Int Urol Nephrol. 2023 Apr;55(4):913-928. doi: 10.1007/s11255-022-03354-7. Epub 2022 Oct 22. Int Urol Nephrol. 2023. PMID: 36271990 Free PMC article. Review.
Cited by
-
Single-cell RNA sequencing in diabetic kidney disease: a literature review.Ren Fail. 2024 Dec;46(2):2387428. doi: 10.1080/0886022X.2024.2387428. Epub 2024 Aug 4. Ren Fail. 2024. PMID: 39099183 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

