Computational identification of long non-coding RNAs associated with graphene therapy in glioblastoma multiforme
- PMID: 38162904
- PMCID: PMC10754320
- DOI: 10.1093/braincomms/fcad293
Computational identification of long non-coding RNAs associated with graphene therapy in glioblastoma multiforme
Abstract
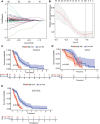
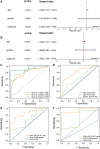
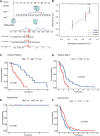
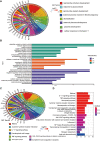
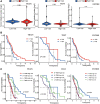
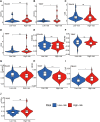
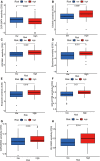
Glioblastoma multiforme represents the most prevalent primary malignant brain tumour, while long non-coding RNA assumes a pivotal role in the pathogenesis and progression of glioblastoma multiforme. Nonetheless, the successful delivery of long non-coding RNA-based therapeutics to the tumour site has encountered significant obstacles attributable to inadequate biocompatibility and inefficient drug delivery systems. In this context, the use of a biofunctional surface modification of graphene oxide has emerged as a promising strategy to surmount these challenges. By changing the surface of graphene oxide, enhanced biocompatibility can be achieved, facilitating efficient transport of long non-coding RNA-based therapeutics specifically to the tumour site. This innovative approach presents the opportunity to exploit the therapeutic potential inherent in long non-coding RNA biology for treating glioblastoma multiforme patients. This study aimed to extract relevant genes from The Cancer Genome Atlas database and associate them with long non-coding RNAs to identify graphene therapy-related long non-coding RNA. We conducted a series of analyses to achieve this goal, including univariate Cox regression, least absolute shrinkage and selection operator regression and multivariate Cox regression. The resulting graphene therapy-related long non-coding RNAs were utilized to develop a risk score model. Subsequently, we conducted Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway analyses on the identified graphene therapy-related long non-coding RNAs. Additionally, we employed the risk model to construct the tumour microenvironment model and analyse drug sensitivity. To validate our findings, we referenced the IMvigor210 immunotherapy model. Finally, we investigated differences in the tumour stemness index. Through our investigation, we identified four promising graphene therapy-related long non-coding RNAs (AC011405.1, HOXC13-AS, LINC01127 and LINC01574) that could be utilized for treating glioblastoma multiforme patients. Furthermore, we identified 16 compounds that could be utilized in graphene therapy. Our study offers novel insights into the treatment of glioblastoma multiforme, and the identified graphene therapy-related long non-coding RNAs and compounds hold promise for further research in this field. Furthermore, additional biological experiments will be essential to validate the clinical significance of our model. These experiments can help confirm the potential therapeutic value and efficacy of the identified graphene therapy-related long non-coding RNAs and compounds in treating glioblastoma multiforme.
Keywords: drug screening; glioblastoma multiforme; graphene oxide therapy; independent prognostic model; lncRNAs.
© The Author(s) 2023. Published by Oxford University Press on behalf of the Guarantors of Brain.
Conflict of interest statement
The authors report no competing interests.
Figures

Similar articles
-
Identification of Novel lncRNA Markers in Glioblastoma Multiforme and Their Clinical Significance: A Study Based on Multiple Sequencing Data.Onco Targets Ther. 2020 Feb 4;13:1087-1098. doi: 10.2147/OTT.S235951. eCollection 2020. Onco Targets Ther. 2020. PMID: 32099410 Free PMC article.
-
Drug-induced modifications and modulations of microRNAs and long non-coding RNAs for future therapy against Glioblastoma Multiforme.Gene. 2020 Jan 10;723:144126. doi: 10.1016/j.gene.2019.144126. Epub 2019 Oct 4. Gene. 2020. PMID: 31589963 Review.
-
Prospective Series of Nine Long Noncoding RNAs Associated with Survival of Patients with Glioblastoma.J Neurol Surg A Cent Eur Neurosurg. 2018 Nov;79(6):471-478. doi: 10.1055/s-0038-1655549. Epub 2018 Jun 29. J Neurol Surg A Cent Eur Neurosurg. 2018. PMID: 29959764
-
Identification of a novel defined inflammation-related long noncoding RNA signature contributes to predicting prognosis and distinction between the cold and hot tumors in bladder cancer.Front Oncol. 2023 Mar 29;13:972558. doi: 10.3389/fonc.2023.972558. eCollection 2023. Front Oncol. 2023. PMID: 37064115 Free PMC article.
-
Exploring miRNA-Associated Signatures with Diagnostic Relevance in Glioblastoma Multiforme and Breast Cancer Patients.J Clin Med. 2015 Aug 14;4(8):1612-30. doi: 10.3390/jcm4081612. J Clin Med. 2015. PMID: 26287251 Free PMC article. Review.
Cited by
-
Combination of transcriptome and Mendelian inheritance reveals novel prognostic biomarker of CTLA-4-related lncRNAs and protective role of nitrogen metabolism pathway in lung adenocarcinoma development.BMC Cancer. 2024 Aug 14;24(1):1009. doi: 10.1186/s12885-024-12777-7. BMC Cancer. 2024. PMID: 39143529 Free PMC article.
-
From vestibular implant to cortex: electrically evoked vestibular responses.J Neurol. 2025 May 29;272(6):430. doi: 10.1007/s00415-025-13158-1. J Neurol. 2025. PMID: 40442486 Free PMC article.
References
-
- Li D, Hu X, Zhang S. Biodegradation of graphene-based nanomaterials in blood plasma affects their biocompatibility, drug delivery, targeted organs and antitumor ability. Biomaterials. 2019;202:12–25. - PubMed
-
- Yang J, He X, Chen L, Zhang Y. Thiol-yne click synthesis of boronic acid functionalized silica nanoparticle-graphene oxide composites for highly selective enrichment of glycoproteins. J Chromatogr A. 2017;1513:118–125. - PubMed
-
- Wang Z, Cheng H, Sheng Y, et al. Biofunctionalized graphene oxide nanosheet for amplifying antitumor therapy: Multimodal high drug encapsulation, prolonged hyperthermal window, and deep-site burst drug release. Biomaterials. 2022;287:121629. - PubMed
-
- Lu W, Yao J, Zhu X, Qi Y. Nanomedicines: Redefining traditional medicine. Biomed Pharmacother. 2020;134:111103. - PubMed
-
- Xu H, Fan M, Elhissi AM, et al. PEGylated graphene oxide for tumor-targeted delivery of paclitaxel. Nanomedicine (Lond). 2015;10(8):1247–1262. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
