Microbial communities of a variety of 75 homemade fermented vegetables
- PMID: 38163080
- PMCID: PMC10757351
- DOI: 10.3389/fmicb.2023.1323424
Microbial communities of a variety of 75 homemade fermented vegetables
Abstract
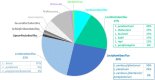
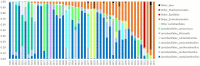
Fermentation is an ancient practice of food preservation. Fermented vegetables are popular in Eastern European and Asian countries. They have received a growing interest in Western countries, where they are mainly manufactured at domestic and artisanal scales and poorly characterized. Our aim was to investigate the microbial communities and the safety of French homemade fermented vegetables, in the frame of a citizen science project. Fermented vegetables and the data associated with their manufacture were collected from citizens and characterized for pH, NaCl concentration, and microbiology by culturomics and 16S DNA metabarcoding analysis. Lactic acid bacteria (LAB) and yeast isolates were identified by 16S rRNA gene sequencing and D1/D2 domains of the large subunit of the rRNA gene, respectively. The 75 collected samples contained 23 types of vegetables, mainly cabbage, followed by carrots and beets, and many mixtures of vegetables. They were 2 weeks to 4 years old, and their median pH was 3.56, except for two samples with a pH over 4.5. LAB represented the dominant viable bacteria. LAB concentrations ranged from non-detectable values to 8.7 log colony-forming units (CFU)/g and only depended on the age of the samples, with the highest most frequently observed in the youngest samples (<100 days). The 93 LAB isolates identified belonged to 23 species, the two mains being Lactiplantibacillus pentosus/plantarum and Levilactobacillus brevis. The other microbial groups enumerated (total aerobic bacteria, halotolerant bacteria, Gram-negative bacteria, and acetic acid bacteria) generally showed lower concentrations compared to LAB concentrations. No pathogenic bacteria were detected. Viable yeasts were observed in nearly half the samples, at concentrations reaching up to 8.0 log CFU/g. The 33 yeast clones identified belonged to 16 species. Bacterial metabarcoding showed two main orders, namely, Lactobacillales (i.e., LAB, 79% of abundance, 177 of the 398 total ASVs) and Enterobacterales (19% of abundance, 191 ASVs). Fifteen LAB genera were identified, with Lactiplantibacillus and Levilactobacillus as the most abundant, with 41 and 12% of total reads, respectively. Enterobacterales members were mainly represented by Enterobacteriaceae and Yersiniaceae. This study is the first wide description of the microbiota of a large variety of homemade fermented vegetables and documents their safety.
Keywords: citizen science; fermented food; lactic acid bacteria; sauerkraut; spontaneous fermentation; yeast.
Copyright © 2023 Thierry, Madec, Chuat, Bage, Picard, Grondin, Rué, Mariadassou, Marché and Valence.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
The lactic acid bacteria and yeast community of home-made sauerkraut from three provinces in Southwest China.Arch Microbiol. 2021 Aug;203(6):3171-3182. doi: 10.1007/s00203-021-02222-9. Epub 2021 Apr 7. Arch Microbiol. 2021. PMID: 33825934
-
Carrot Juice Fermentations as Man-Made Microbial Ecosystems Dominated by Lactic Acid Bacteria.Appl Environ Microbiol. 2018 May 31;84(12):e00134-18. doi: 10.1128/AEM.00134-18. Print 2018 Jun 15. Appl Environ Microbiol. 2018. PMID: 29654180 Free PMC article.
-
The microbiota of Lafun, an African traditional cassava food product.Int J Food Microbiol. 2009 Jul 31;133(1-2):22-30. doi: 10.1016/j.ijfoodmicro.2009.04.019. Epub 2009 Apr 24. Int J Food Microbiol. 2009. PMID: 19493582
-
A Holistic Review on Euro-Asian Lactic Acid Bacteria Fermented Cereals and Vegetables.Microorganisms. 2020 Aug 3;8(8):1176. doi: 10.3390/microorganisms8081176. Microorganisms. 2020. PMID: 32756333 Free PMC article. Review.
-
Lactic acid bacteria from fermented table olives.Food Microbiol. 2012 Aug;31(1):1-8. doi: 10.1016/j.fm.2012.01.006. Epub 2012 Feb 2. Food Microbiol. 2012. PMID: 22475936 Review.
Cited by
-
Physicochemical, Nutritional, and Antioxidant Properties of Traditionally Fermented Thai Vegetables: A Promising Functional Plant-Based Food.Foods. 2024 Sep 8;13(17):2848. doi: 10.3390/foods13172848. Foods. 2024. PMID: 39272613 Free PMC article.
-
Improving the bacterial community, flavor, and safety properties of northeastern sauerkraut by inoculating autochthonous Levilactobacillus brevis.Food Chem X. 2024 Apr 24;22:101408. doi: 10.1016/j.fochx.2024.101408. eCollection 2024 Jun 30. Food Chem X. 2024. PMID: 38707785 Free PMC article.
-
Sustainable Innovations in Food Microbiology: Fermentation, Biocontrol, and Functional Foods.Foods. 2025 Jun 30;14(13):2320. doi: 10.3390/foods14132320. Foods. 2025. PMID: 40647071 Free PMC article. Review.
-
Spontaneously fermented Allium cepa L. as a source of lactic acid bacteria with probiotic potential.Sci Rep. 2025 Jul 17;15(1):25970. doi: 10.1038/s41598-025-10037-7. Sci Rep. 2025. PMID: 40676012 Free PMC article.
-
The Short- and Long-Term Effects of a Short Course of Sauerkraut Supplementation on the Gut Microbiota of Active Athletes: A Pilot Follow-Up Study.Nutrients. 2025 Mar 6;17(5):929. doi: 10.3390/nu17050929. Nutrients. 2025. PMID: 40077799 Free PMC article.
References
-
- Ballester E., Ribes S., Barat J. M., Fuentes A. (2022). Spoilage yeasts in fermented vegetables: conventional and novel control strategies. Eur. Food Res. Technol. 248, 315–328. doi: 10.1007/s00217-021-03888-7 - DOI
-
- Beckers B., Op De Beeck M., Thijs S., Truyens S., Weyens N., Boerjan W., et al. . (2016). Performance of 16s rDNA primer pairs in the study of rhizosphere and Endosphere bacterial microbiomes in metabarcoding studies. Front. Microbiol. 7:650. doi: 10.3389/fmicb.2016.00650, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

