Transcription of biochemical defenses by the harmful brown tide pelagophyte, Aureococcus anophagefferens, in response to the protozoan grazer, Oxyrrhis marina
- PMID: 38163083
- PMCID: PMC10756674
- DOI: 10.3389/fmicb.2023.1295160
Transcription of biochemical defenses by the harmful brown tide pelagophyte, Aureococcus anophagefferens, in response to the protozoan grazer, Oxyrrhis marina
Abstract
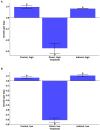
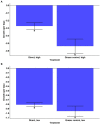
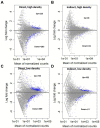
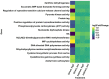
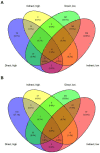
Aureococcus anophagefferens is a small marine pelagophyte that forms recurrent harmful brown tides blooms with adverse ecological and economic impacts. During blooms, A. anophagefferens experiences lower zooplankton grazing mortality than other phytoplankton potentially due to the synthesis of anti-predator compounds including extracellular polysaccharides. This study characterized the transcriptomic response of A. anophagefferens when exposed to the protozooplankton, Oxyrrhis marina, and assessed whether this response involved chemical cues. Transcriptomes were generated from A. anophagefferens populations grown at high (1×106 cells mL-1) and low (5×105 cells mL-1) cell densities incubated directly with O. marina or receiving only filtrate from co-cultures of A. anophagefferens and O. marina to evaluate the role of chemical cues. There were a greater number of genes differentially expressed in response to grazing in the lower concentration of A. anophagefferens compared to the high concentration treatment and in response to direct grazing compared to filtrate. KEGG pathway analysis revealed that direct grazer exposure led to a significant increase in transcripts of genes encoding secondary metabolite production (p < 0.001). There was broad transcriptional evidence indicating the induction of biosynthetic pathways for polyketides and sterols in response to zooplankton grazers, compounds associated with damage to marine organisms. In addition, exposure to O. marina elicited changes in the abundance of transcripts associated with carbohydrate metabolism that could support the formation of an extracellular polysaccharide matrix including genes related to glycoprotein synthesis and carbohydrate transport. Collectively, these findings support the hypothesis that A. anophagefferens can induce biochemical pathways that reduce grazing mortality and support blooms.
Keywords: Brown tide; gene express; harmful algal bloom; induced defense; zooplankton grazing.
Copyright © 2023 Dawydiak and Gobler.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Infection by a Giant Virus (AaV) Induces Widespread Physiological Reprogramming in Aureococcus anophagefferens CCMP1984 - A Harmful Bloom Algae.Front Microbiol. 2018 Apr 19;9:752. doi: 10.3389/fmicb.2018.00752. eCollection 2018. Front Microbiol. 2018. PMID: 29725322 Free PMC article.
-
Green Tides in the Yellow Sea Promoted the Proliferation of Pelagophyte Aureococcus anophagefferens.Environ Sci Technol. 2022 Mar 1;56(5):3056-3064. doi: 10.1021/acs.est.1c06502. Epub 2022 Feb 8. Environ Sci Technol. 2022. PMID: 35133807
-
Relationship between phytoplankton community succession and environmental parameters in Qinhuangdao coastal areas, China: A region with recurrent brown tide outbreaks.Ecotoxicol Environ Saf. 2018 Sep 15;159:85-93. doi: 10.1016/j.ecoenv.2018.04.043. Epub 2018 May 3. Ecotoxicol Environ Saf. 2018. PMID: 29730413
-
Kratosvirus quantuckense: the history and novelty of an algal bloom disrupting virus and a model for giant virus research.Front Microbiol. 2023 Nov 30;14:1284617. doi: 10.3389/fmicb.2023.1284617. eCollection 2023. Front Microbiol. 2023. PMID: 38098665 Free PMC article. Review.
-
Marine harmful algal blooms (HABs) in the United States: History, current status and future trends.Harmful Algae. 2021 Feb;102:101975. doi: 10.1016/j.hal.2021.101975. Epub 2021 Mar 3. Harmful Algae. 2021. PMID: 33875183 Free PMC article. Review.
References
-
- Andrews S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data [Online]. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
-
- Bendif E. M., Probert I., Schroeder D. C., de Vargas C. (2013). On the description of Tisochrysis lutea gen. Nov. Sp. Nov. And Isochrysis nuda sp. Nov. In the Isochrysidales, and the transfer of Dicrateria to the Prymnesiales (Haptophyta). J. Appl. Phycol. 25, 1763–1776. doi: 10.1007/s10811-013-0037-0 - DOI
LinkOut - more resources
Full Text Sources

