Novel molecular classification and prognosis of papillary renal cell carcinoma based on a large-scale CRISPR-Cas9 screening and machine learning
- PMID: 38163209
- PMCID: PMC10754875
- DOI: 10.1016/j.heliyon.2023.e23184
Novel molecular classification and prognosis of papillary renal cell carcinoma based on a large-scale CRISPR-Cas9 screening and machine learning
Abstract
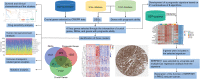
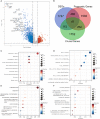
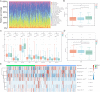
Papillary renal cell carcinoma (PRCC) is a highly heterogeneous cancer, and PRCC patients with advanced/metastatic subgroup showed obviously shorter survival compared to other kinds of renal cell carcinomas. However, the molecular mechanism and prognostic predictors of PRCC remain unclear and are worth deep studying. The aim of this study is to identify novel molecular classification and construct a reliable prognostic model for PRCC. The expression data were retrieved from TCGA, GEO, GTEx and TARGET databases. CRISPR data was obtained from Depmap database. The key genes were selected by the intersection of CRISPR-Cas9 screening genes, differentially expressed genes, and genes with prognostic capacity in PRCC. The molecular classification was identified based on the key genes. Drug sensitivity, tumor microenvironment, somatic mutation, and survival were compared among the novel classification. A prognostic model utilizing multiple machine learning algorithms based on the key genes was developed and tested by independent external validation set. Our study identified three clusters (C1, C2 and C3) in PRCC based on 41 key genes. C2 had obviously higher expression of the key genes and lower survival than C1 and C3. Significant differences in drug sensitivity, tumor microenvironment, and mutation landscape have been observed among the three clusters. By utilizing 21 combinations of 9 machine learning algorithms, 9 out of 41 genes were chosen to construct a robust prognostic signature, which exhibited good prognostic ability. SERPINH1 was identified as a critical gene for its strong prognostic ability in PRCC by univariate and multiple Cox regression analyses. Quantitative real-time PCR and Western blot demonstrated that SERPINH1 mRNA and protein were highly expressed in PRCC cells compared with normal human renal cells. This study exhibited a new molecular classification and prognostic signature for PRCC, which may provide a potential biomarker and therapy target for PRCC patients.
Keywords: CRISPR-Cas9; Depmap; Machine learning; Papillary renal cell carcinoma; SERPINH1.
© 2023 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures








References
LinkOut - more resources
Full Text Sources
Miscellaneous

