DCAF13 inhibits the p53 signaling pathway by promoting p53 ubiquitination modification in lung adenocarcinoma
- PMID: 38163876
- PMCID: PMC10759521
- DOI: 10.1186/s13046-023-02936-2
DCAF13 inhibits the p53 signaling pathway by promoting p53 ubiquitination modification in lung adenocarcinoma
Abstract
Background: Lung cancer is a malignant tumor with the highest mortality worldwide. Abnormalities in the ubiquitin proteasome system are considered to be contributed to lung cancer progression with deleterious effects. DDB1 and CUL4 associated factor 13 (DCAF13) is a substrate receptor of the E3 ubiquitin ligase CRL4, but its role in lung cancer remains unknown. In this study, we aimed to investigate the regulatory mechanisms of DCAF13 in lung adenocarcinoma (LUAD).
Methods: So as to investigate the effect of DCAF13 on lung adenocarcinoma cell function using in vivo and in vitro. Mechanistically, we have identified the downstream targets of DCAF13 by using RNA-sequencing, as well as ubiquitination assays, co-immunoprecipitation, immunofluorescence, immunohistochemistry and chromatin immunoprecipitation - qPCR experiments.
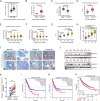
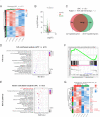
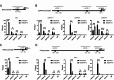
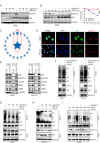
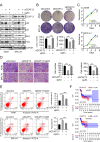
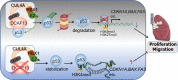
Results: Our findings reveal that DCAF13 is a carcinogenic factor in LUAD, as it is highly expressed and negatively correlated with clinical outcomes in LUAD patients. Through RNA-sequencing, it has been shown that DCAF13 negatively regulates the p53 signaling pathway and inhibits p53 downstream targets including p21, BAX, FAS, and PIDD1. We also demonstrate that DCAF13 can bind to p53 protein, leading to K48-linked ubiquitination and degradation of p53. Functionally, we have shown that DCAF13 knockdown inhibits cell proliferation and migration. Our results highlight the significant role of DCAF13 in promoting LUAD progression by inhibiting p53 protein stabilization and the p53 signaling pathway. Furthermore, our findings suggest that high DCAF13 expression is a poor prognostic indicator in LUAD, and DCAF13 may be a potential therapeutic target for treating with this aggressive cancer.
Conclusions: The DCAF13 as a novel negative regulator of p53 to promote LUAD progression via facilitating p53 ubiquitination and degradation, suggesting that DCAF13 might be a novel biomarker and therapeutical target for LUAD.
Keywords: DDB1 and CUL4 associated factor 13; Lung adenocarcinoma; Protein degradation; Ubiquitination; p53 signaling pathway.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Sung H, Ferlay J, Siegel RL et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin, 2021;71(3):209–249. - PubMed
MeSH terms
Substances
Grants and funding
- 2023J387/Ningbo Natural Science Foundation
- 2024KY373/Medical Science and Technology Project of Zhejiang Province
- 2023KY303/Medical Science and Technology Project of Zhejiang Province
- 2023RC270/Medical Science and Technology Project of Zhejiang Province
- LQ20H160010/Natural Science Foundation of Zhejiang Province
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

