Sarcosine dehydrogenase as an immune infiltration-associated biomarker for the prognosis of hepatocellular carcinoma
- PMID: 38164283
- PMCID: PMC10751682
- DOI: 10.7150/jca.89616
Sarcosine dehydrogenase as an immune infiltration-associated biomarker for the prognosis of hepatocellular carcinoma
Abstract
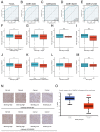
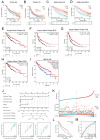
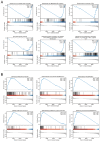
This study was aimed to investigate the prognostic value and clinical significance of sarcosine dehydrogenase (SARDH) in hepatocellular carcinoma (HCC) and to explore the underlying mechanisms. The Cancer Genome Atlas (TCGA), Gene Expression Omnibus (GEO), HPA and CPTAC databases were adopted to analyze the expression of SARDH mRNA and protein between normal liver tissue and HCC, and examine their relationship with clinicopathological features. Kaplan-Meier analysis, Cox regression, as well as nomogram were adopted to explore the prognostic value of SARDH in HCC. Gene Ontology (GO), Kyoto Gene and Genome Encyclopedia (KEGG) together with Gene Set Enrichment Analysis (GSEA) were adopted to analyze the molecular mechanisms and biological functions of SARDH in HCC; while MethSurv, STRING, GeneMANIA, TIMER database data and single-sample gene set enrichment analysis (ssGSEA) algorithm were used for other bioinformatic analysis. Furthermore, immunohistochemistry was used to verify the expression of SARDH. Compared to normal liver tissue, SARDH expression was markedly lower in HCC. A lower SARDH expression was linked with Pathologic T stage (T3&T4), pathologic stage (Stage III&IV), and histologic grade (G3&4), which further indicates worse prognosis. Besides, results of bioinformatic analysis proved that SARDH expression was correlated with immune infiltration. In addition, SARDH hypermethylation was related to a poorer prognosis. SARDH expression was related to several key genes in the Ferroptosis pathway.
Keywords: Biomarkers; Hepatocellular carcinoma; Immune infiltration; Prognostic; Sarcosine dehydrogenase.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures





References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: a cancer journal for clinicians. 2021;71:209–49. - PubMed
-
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a cancer journal for clinicians. 2018;68:394–424. - PubMed
-
- Li X, Ramadori P, Pfister D, Seehawer M, Zender L, Heikenwalder M. The immunological and metabolic landscape in primary and metastatic liver cancer. Nat Rev Cancer. 2021;21:541–57. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

