Genomic characterization of Salmonella isolated from retail chicken and humans with diarrhea in Qingdao, China
- PMID: 38164401
- PMCID: PMC10757937
- DOI: 10.3389/fmicb.2023.1295769
Genomic characterization of Salmonella isolated from retail chicken and humans with diarrhea in Qingdao, China
Abstract
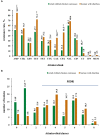
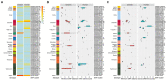
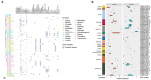
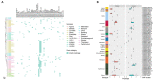
Salmonella, especially antimicrobial resistant strains, remains one of the leading causes of foodborne bacterial disease. Retail chicken is a major source of human salmonellosis. Here, we investigated the prevalence, antimicrobial resistance (AMR), and genomic characteristics of Salmonella in 88 out of 360 (24.4%) chilled chicken carcasses, together with 86 Salmonella from humans with diarrhea in Qingdao, China in 2020. The most common serotypes were Enteritidis and Typhimurium (including the serotype I 4,[5],12:i:-) among Salmonella from both chicken and humans. The sequence types were consistent with serotypes, with ST11, ST34 and ST19 the most dominantly identified. Resistance to nalidixic acid, ampicillin, tetracycline and chloramphenicol were the top four detected in Salmonella from both chicken and human sources. High multi-drug resistance (MDR) and resistance to third-generation cephalosporins resistance were found in Salmonella from chicken (53.4%) and humans (75.6%). In total, 149 of 174 (85.6%) Salmonella isolates could be categorized into 60 known SNP clusters, with 8 SNP clusters detected in both sources. Furthermore, high prevalence of plasmid replicons and prophages were observed among the studied isolates. A total of 79 antimicrobial resistant genes (ARGs) were found, with aac(6')-Iaa, blaTEM-1B, tet(A), aph(6)-Id, aph(3″)-Ib, sul2, floR and qnrS1 being the dominant ARGs. Moreover, nine CTX-M-type ESBL genes and the genes blaNMD-1, mcr-1.1, and mcr-9.1 were detected. The high incidence of MDR Salmonella, especially possessing lots of mobile genetic elements (MGEs) in this study posed a severe risk to food safety and public health, highlighting the importance of improving food hygiene measures to reduce the contamination and transmission of this bacterium. Overall, it is essential to continue monitoring the Salmonella serotypes, implement the necessary prevention and strategic control plans, and conduct an epidemiological surveillance system based on whole-genome sequencing.
Keywords: Salmonella; antimicrobial resistance; genome sequencing; humans with diarrhea; retail chicken.
Copyright © 2023 Wang, Cui, Liu, Hu, Li, Zhou, Deng, Dong, Li and Xiao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Characterization of the prevalence of Salmonella in different retail chicken supply modes using genome-wide and machine-learning analyses.Food Res Int. 2024 Sep;191:114654. doi: 10.1016/j.foodres.2024.114654. Epub 2024 Jun 19. Food Res Int. 2024. PMID: 39059904
-
Genomic Characterization of Antimicrobial-Resistant Salmonella enterica in Duck, Chicken, and Pig Farms and Retail Markets in Eastern China.Microbiol Spectr. 2022 Oct 26;10(5):e0125722. doi: 10.1128/spectrum.01257-22. Epub 2022 Sep 1. Microbiol Spectr. 2022. PMID: 36047803 Free PMC article.
-
Antimicrobial resistance and resistance genes in Salmonella strains isolated from broiler chickens along the slaughtering process in China.Int J Food Microbiol. 2017 Oct 16;259:43-51. doi: 10.1016/j.ijfoodmicro.2017.07.023. Epub 2017 Aug 1. Int J Food Microbiol. 2017. PMID: 28800411
-
Antimicrobial resistance and genomic investigation of non-typhoidal Salmonella isolated from outpatients in Shaoxing city, China.Front Public Health. 2022 Sep 13;10:988317. doi: 10.3389/fpubh.2022.988317. eCollection 2022. Front Public Health. 2022. PMID: 36176509 Free PMC article.
-
Prevalence and antimicrobial resistance of retail-meat-borne Salmonella in southern China during the years 2009-2016: The diversity of contamination and the resistance evolution of multidrug-resistant isolates.Int J Food Microbiol. 2020 Nov 16;333:108790. doi: 10.1016/j.ijfoodmicro.2020.108790. Epub 2020 Jul 14. Int J Food Microbiol. 2020. PMID: 32693316
Cited by
-
Nationwide surveillance and characterization of the third-generation cephalosporin-resistant Salmonella enterica serovar infantis isolated from chickens in South Korea between 2010 and 2022.Heliyon. 2024 Aug 29;10(17):e37124. doi: 10.1016/j.heliyon.2024.e37124. eCollection 2024 Sep 15. Heliyon. 2024. PMID: 39319126 Free PMC article.
-
Whole-genome sequencing analysis of Salmonella enterica serotype Enteritidis isolated from poultry sources in Mongolia.Front Vet Sci. 2025 May 20;12:1595674. doi: 10.3389/fvets.2025.1595674. eCollection 2025. Front Vet Sci. 2025. PMID: 40463800 Free PMC article.
-
Prevalence and transmission of Salmonella collected from farming to egg processing of layer production chain in Jiangsu Province, China.Poult Sci. 2025 Feb;104(2):104714. doi: 10.1016/j.psj.2024.104714. Epub 2024 Dec 20. Poult Sci. 2025. PMID: 39721277 Free PMC article.
References
LinkOut - more resources
Full Text Sources

