Signaling Specificity and Kinetics of the Human Metabotropic Glutamate Receptors
- PMID: 38164584
- PMCID: PMC10794986
- DOI: 10.1124/molpharm.123.000795
Signaling Specificity and Kinetics of the Human Metabotropic Glutamate Receptors
Abstract
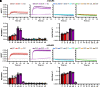
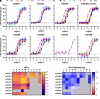
Metabotropic glutamate receptors (mGluRs) are obligate dimer G protein coupled receptors that can all function as homodimers. Here, each mGluR homodimer was examined for its G protein coupling profile using a bioluminescence resonance energy transfer-based assay that detects the interaction between a split YFP-tagged Gβ 1γ2 and a Nanoluciferase tagged free Gβγ sensor, MAS-GRK3-ct- nanoluciferase with 14 specific Gα proteins heterologously expressed, representing each family. Canonically, the group II and III mGluRs (2 and 3 and 4, 6, 7, and 8, respectively) are thought to couple to Gi/o exclusively. In addition, the group I mGluRs (1 and 5) are known to couple to the Gq/11 family and generally thought to also couple to the pertussis toxin-sensitive Gi/o family some reports have suggested Gs coupling is possible as cAMP elevations have been noted. In this study, coupling was observed with all eight mGluRs through the Gi/o proteins and only mGluR1 and mGluR5 through Gq/11, and, perhaps surprisingly, not G14 None activated any Gs protein. Interestingly, coupling was seen with the group I and II but not the group III mGluRs to G16 Slow but significant coupling to Gz was also seen with the group II receptors. SIGNIFICANCE STATEMENT: Metabotropic glutamate receptor (mGluR)-G protein coupling has not been thoroughly examined, and some controversy remains about whether some mGluRs can activate Gαs family members. Here we examine the ability of each mGluR to activate representative members of every Gα protein family. While all mGluRs can activate Gαi/o proteins, only the group I mGluRs couple to Gαq/11, and no members of the family can activate Gαs family members, including the group I receptors alone or with positive allosteric modulators.
Copyright © 2024 by The American Society for Pharmacology and Experimental Therapeutics.
Figures





Update of
-
Signaling specificity and kinetics of the human metabotropic glutamate receptors.bioRxiv [Preprint]. 2023 Jul 28:2023.07.24.550373. doi: 10.1101/2023.07.24.550373. bioRxiv. 2023. Update in: Mol Pharmacol. 2024 Jan 10;105(2):104-115. doi: 10.1124/molpharm.123.000795. PMID: 37546908 Free PMC article. Updated. Preprint.
References
-
- Choi JJ, Nam KH, Min B, Kim SJ, Söll D, Kwon ST (2006) Protein trans-splicing and characterization of a split family B-type DNA polymerase from the hyperthermophilic archaeal parasite Nanoarchaeum equitans. J Mol Biol 356:1093–1106. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
