Nanobody-Mediated Dualsteric Engagement of the Angiotensin Receptor Broadens Biased Ligand Pharmacology
- PMID: 38164609
- PMCID: PMC10877709
- DOI: 10.1124/molpharm.123.000797
Nanobody-Mediated Dualsteric Engagement of the Angiotensin Receptor Broadens Biased Ligand Pharmacology
Abstract
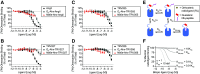
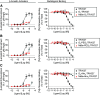
Dualsteric G protein-coupled receptor (GPCR) ligands are a class of bitopic ligands that consist of an orthosteric pharmacophore, which binds to the pocket occupied by the receptor's endogenous agonist, and an allosteric pharmacophore, which binds to a distinct site. These ligands have the potential to display characteristics of both orthosteric and allosteric ligands. To explore the signaling profiles that dualsteric ligands of the angiotensin II type 1 receptor (AT1R) can access, we ligated a 6e epitope tag-specific nanobody (single-domain antibody fragment) to angiotensin II (AngII) and analogs that show preferential allosteric coupling to Gq (TRV055, TRV056) or β-arrestin (TRV027). While the nanobody itself acts as a probe-specific neutral or negative allosteric ligand of N-terminally 6e-tagged AT1R, nanobody conjugation to orthosteric ligands had varying effects on Gq dissociation and β-arrestin plasma membrane recruitment. The potency of certain AngII analogs was enhanced up to 100-fold, and some conjugates behaved as partial agonists, with up to a 5-fold decrease in maximal efficacy. Nanobody conjugation also biased the signaling of TRV055 and TRV056 toward Gq, suggesting that Gq bias at AT1R can be modulated through molecular mechanisms distinct from those previously elucidated. Both competition radioligand binding experiments and functional assays demonstrated that orthosteric antagonists (angiotensin receptor blockers) act as non-competitive inhibitors of all these nanobody-peptide conjugates. This proof-of-principle study illustrates the array of pharmacological patterns that can be achieved by incorporating neutral or negative allosteric pharmacophores into dualsteric ligands. Nanobodies directed toward linear epitopes could provide a rich source of allosteric reagents for this purpose. SIGNIFICANCE STATEMENT: Here we engineer bitopic (dualsteric) ligands for epitope-tagged angiotensin II type 1 receptor by conjugating angiotensin II or its biased analogs to an epitope-specific nanobody (antibody fragment). Our data demonstrate that nanobody-mediated interactions with the receptor N-terminus endow angiotensin analogs with properties of allosteric modulators and provide a novel mechanism to increase the potency, modulate the maximal effect, or alter the bias of ligands.
U.S. Government work not protected by U.S. copyright.
Figures
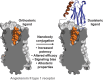





References
-
- Antony J, Kellershohn K, Mohr-Andrä M, Kebig A, Prilla S, Muth M, Heller E, Disingrini T, Dallanoce C, Bertoni S, et al. (2009) Dualsteric GPCR targeting: a novel route to binding and signaling pathway selectivity. FASEB J 23:442–450. - PubMed
-
- Ballesteros JA, Weinstein H (1995) Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors, in Methods in Neurosciences (Sealfon SC, ed) pp 366–428, Academic Press.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
