Heterogeneous nuclear ribonucleoprotein K promotes cap-independent translation initiation of retroviral mRNAs
- PMID: 38165048
- PMCID: PMC10954487
- DOI: 10.1093/nar/gkad1221
Heterogeneous nuclear ribonucleoprotein K promotes cap-independent translation initiation of retroviral mRNAs
Abstract
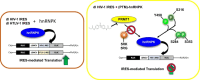
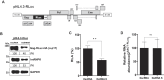
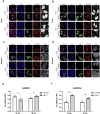
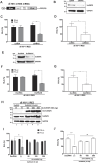
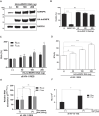
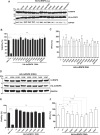
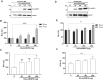
Translation initiation of the human immunodeficiency virus-type 1 (HIV-1) genomic mRNA (vRNA) is cap-dependent or mediated by an internal ribosome entry site (IRES). The HIV-1 IRES requires IRES-transacting factors (ITAFs) for function. In this study, we evaluated the role of the heterogeneous nuclear ribonucleoprotein K (hnRNPK) as a potential ITAF for the HIV-1 IRES. In HIV-1-expressing cells, the depletion of hnRNPK reduced HIV-1 vRNA translation. Furthermore, both the depletion and overexpression of hnRNPK modulated HIV-1 IRES activity. Phosphorylations and protein arginine methyltransferase 1 (PRMT1)-induced asymmetrical dimethylation (aDMA) of hnRNPK strongly impacted the protein's ability to promote the activity of the HIV-1 IRES. We also show that hnRNPK acts as an ITAF for the human T cell lymphotropic virus-type 1 (HTLV-1) IRES, present in the 5'UTR of the viral sense mRNA, but not for the IRES present in the antisense spliced transcript encoding the HTLV-1 basic leucine zipper protein (sHBZ). This study provides evidence for a novel role of the host hnRNPK as an ITAF that stimulates IRES-mediated translation initiation for the retroviruses HIV-1 and HTLV-1.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




Similar articles
-
The Mytilus chilensis Steamer-like Element-1 Retrotransposon Antisense mRNA Harbors an Internal Ribosome Entry Site That Is Modulated by hnRNPK.Viruses. 2024 Mar 5;16(3):403. doi: 10.3390/v16030403. Viruses. 2024. PMID: 38543768 Free PMC article.
-
Post-translational modifications of hnRNP A1 differentially modulate retroviral IRES-mediated translation initiation.Nucleic Acids Res. 2020 Oct 9;48(18):10479-10499. doi: 10.1093/nar/gkaa765. Nucleic Acids Res. 2020. PMID: 32960212 Free PMC article.
-
hnRNP K Is a Novel Internal Ribosomal Entry Site-Transacting Factor That Negatively Regulates Foot-and-Mouth Disease Virus Translation and Replication and Is Antagonized by Viral 3C Protease.J Virol. 2020 Aug 17;94(17):e00803-20. doi: 10.1128/JVI.00803-20. Print 2020 Aug 17. J Virol. 2020. PMID: 32581104 Free PMC article.
-
Hepatitis C Virus Translation Regulation.Int J Mol Sci. 2020 Mar 27;21(7):2328. doi: 10.3390/ijms21072328. Int J Mol Sci. 2020. PMID: 32230899 Free PMC article. Review.
-
Elusive Trans-Acting Factors Which Operate with Type I (Poliovirus-like) IRES Elements.Int J Mol Sci. 2022 Dec 7;23(24):15497. doi: 10.3390/ijms232415497. Int J Mol Sci. 2022. PMID: 36555135 Free PMC article. Review.
Cited by
-
Targeting WDPF domain of Hsp27 achieves a broad spectrum of antiviral.MedComm (2020). 2025 Feb 26;6(3):e70032. doi: 10.1002/mco2.70032. eCollection 2025 Mar. MedComm (2020). 2025. PMID: 40013315 Free PMC article.
-
Heterogeneous Ribonucleoprotein K Is a Host Regulatory Factor of Chikungunya Virus Replication in Astrocytes.Viruses. 2024 Dec 14;16(12):1918. doi: 10.3390/v16121918. Viruses. 2024. PMID: 39772225 Free PMC article.
-
Host RNA-Binding Proteins as Regulators of HIV-1 Replication.Viruses. 2024 Dec 31;17(1):43. doi: 10.3390/v17010043. Viruses. 2024. PMID: 39861832 Free PMC article. Review.
-
Interferon-Regulated Expression of Cellular Splicing Factors Modulates Multiple Levels of HIV-1 Gene Expression and Replication.Viruses. 2024 Jun 11;16(6):938. doi: 10.3390/v16060938. Viruses. 2024. PMID: 38932230 Free PMC article. Review.
-
The Mytilus chilensis Steamer-like Element-1 Retrotransposon Antisense mRNA Harbors an Internal Ribosome Entry Site That Is Modulated by hnRNPK.Viruses. 2024 Mar 5;16(3):403. doi: 10.3390/v16030403. Viruses. 2024. PMID: 38543768 Free PMC article.
References
-
- Barrera A., Olguin V., Vera-Otarola J., Lopez-Lastra M. Cap-independent translation initiation of the unspliced RNA of retroviruses. Biochim. Biophys. Acta Gene Regul. Mech. 2020; 1863:194583. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

