Comprehensive characterization of coding and non-coding single nucleotide polymorphisms of the Myoneurin (MYNN) gene using molecular dynamics simulation and docking approaches
- PMID: 38165846
- PMCID: PMC10760682
- DOI: 10.1371/journal.pone.0296361
Comprehensive characterization of coding and non-coding single nucleotide polymorphisms of the Myoneurin (MYNN) gene using molecular dynamics simulation and docking approaches
Abstract
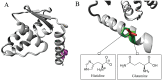
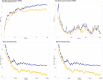
Genome-wide association studies (GWAS) identified a coding single nucleotide polymorphism, MYNN rs10936599, at chromosome 3q. MYNN gene encodes myoneurin protein, which has been associated with several cancer pathogenesis and disease development processes. However, there needed to be a more detailed characterization of this polymorphism's (and other coding and non-coding polymorphisms) structural, functional, and molecular impact. The current study addressed this gap and analyzed different properties of rs10936599 and non-coding SNPs of MYNN via a thorough computational method. The variant, rs10936599, was predicted functionally deleterious by nine functionality prediction approaches, like SIFT, PolyPhen-2, and REVEL, etc. Following that, structural modifications were estimated through the HOPE server and Mutation3D. Moreover, the mutation was found in a conserved and active residue, according to ConSurf and CPORT. Further, the secondary structures were predicted, followed by tertiary structures, and there was a significant deviation between the native and variant models. Similarly, molecular simulation also showed considerable differences in the dynamic pattern of the wildtype and mutant structures. Molecular docking revealed that the variant binds with better docking scores with ligand NOTCH2. In addition to that, non-coding SNPs located at the MYNN locus were retrieved from the ENSEMBL database. These were found to disrupt the transcription factor binding regulatory regions; nonetheless, only two affect miRNA target sites. Again, eight non-coding variants were detected in the testes with normalized expression, whereas HaploReg v4.1 unveiled annotations for non-coding variants. In summary, in silico comprehensive characterization of coding and non-coding single nucleotide polymorphisms of MYNN gene will assist researchers to work on MYNN gene and establish their association with certain types of cancers.
Copyright: © 2024 Mou et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors confirm that they have no known conflict of interest that could be considered to have influenced the research presented in this study.
Figures






Similar articles
-
Comprehensive characterization of high-risk coding and non-coding single nucleotide polymorphisms of human CXCR4 gene.PLoS One. 2024 Dec 23;19(12):e0312733. doi: 10.1371/journal.pone.0312733. eCollection 2024. PLoS One. 2024. PMID: 39715225 Free PMC article.
-
Comprehensive Characterization of the Coding and Non-Coding Single Nucleotide Polymorphisms in the Tumor Protein p63 (TP63) Gene Using In Silico Tools.Biomolecules. 2021 Nov 20;11(11):1733. doi: 10.3390/biom11111733. Biomolecules. 2021. PMID: 34827731 Free PMC article.
-
The Association of MYNN and TERC Gene Polymorphisms and Bladder Cancer in a Turkish Population.Urol J. 2019 Feb 21;16(1):50-55. doi: 10.22037/uj.v0i0.4083. Urol J. 2019. PMID: 30120764
-
Variation in the Untranslated Genome and Susceptibility to Infections.Front Immunol. 2018 Sep 7;9:2046. doi: 10.3389/fimmu.2018.02046. eCollection 2018. Front Immunol. 2018. PMID: 30245696 Free PMC article. Review.
-
Making sense of GWAS: using epigenomics and genome engineering to understand the functional relevance of SNPs in non-coding regions of the human genome.Epigenetics Chromatin. 2015 Dec 30;8:57. doi: 10.1186/s13072-015-0050-4. eCollection 2015. Epigenetics Chromatin. 2015. PMID: 26719772 Free PMC article. Review.
Cited by
-
Comprehensive characterization of high-risk coding and non-coding single nucleotide polymorphisms of human CXCR4 gene.PLoS One. 2024 Dec 23;19(12):e0312733. doi: 10.1371/journal.pone.0312733. eCollection 2024. PLoS One. 2024. PMID: 39715225 Free PMC article.
-
The co-location of CD14+APOE+ cells and MMP7+ tumour cells contributed to worse immunotherapy response in non-small cell lung cancer.Clin Transl Med. 2024 Sep;14(9):e70009. doi: 10.1002/ctm2.70009. Clin Transl Med. 2024. PMID: 39187937 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

