Multi-omic approaches for host-microbiome data integration
- PMID: 38166610
- PMCID: PMC10766395
- DOI: 10.1080/19490976.2023.2297860
Multi-omic approaches for host-microbiome data integration
Abstract
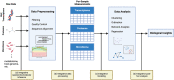
The gut microbiome interacts with the host through complex networks that affect physiology and health outcomes. It is becoming clear that these interactions can be measured across many different omics layers, including the genome, transcriptome, epigenome, metabolome, and proteome, among others. Multi-omic studies of the microbiome can provide insight into the mechanisms underlying host-microbe interactions. As more omics layers are considered, increasingly sophisticated statistical methods are required to integrate them. In this review, we provide an overview of approaches currently used to characterize multi-omic interactions between host and microbiome data. While a large number of studies have generated a deeper understanding of host-microbiome interactions, there is still a need for standardization across approaches. Furthermore, microbiome studies would also benefit from the collection and curation of large, publicly available multi-omics datasets.
Keywords: Multiomics; analysis; disease; host-microbiome interactions; inference; microbiome; network.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

Similar articles
-
A framework for predictive modeling of microbiome multi-omics data: latent interacting variable-effects (LIVE) modeling.BMC Bioinformatics. 2025 Apr 29;26(1):115. doi: 10.1186/s12859-025-06134-z. BMC Bioinformatics. 2025. PMID: 40301728 Free PMC article.
-
Network of Interactions Between Gut Microbiome, Host Biomarkers, and Urine Metabolome in Carotid Atherosclerosis.Front Cell Infect Microbiol. 2021 Oct 7;11:708088. doi: 10.3389/fcimb.2021.708088. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34692558 Free PMC article.
-
Unfolding and de-confounding: biologically meaningful causal inference from longitudinal multi-omic networks using METALICA.mSystems. 2024 Oct 22;9(10):e0130323. doi: 10.1128/msystems.01303-23. Epub 2024 Sep 6. mSystems. 2024. PMID: 39240096 Free PMC article.
-
Multi-omics insights into the interplay between gut microbiota and colorectal cancer in the "microworld" age.Mol Omics. 2023 May 9;19(4):283-296. doi: 10.1039/d2mo00288d. Mol Omics. 2023. PMID: 36916422 Review.
-
Host-microbe interactions: Profiles in the transcriptome, the proteome, and the metabolome.Periodontol 2000. 2020 Feb;82(1):115-128. doi: 10.1111/prd.12316. Periodontol 2000. 2020. PMID: 31850641 Free PMC article. Review.
Cited by
-
Integrating the milk microbiome signatures in mastitis: milk-omics and functional implications.World J Microbiol Biotechnol. 2025 Jan 18;41(2):41. doi: 10.1007/s11274-024-04242-1. World J Microbiol Biotechnol. 2025. PMID: 39826029 Free PMC article. Review.
-
Identifying Optimal Machine Learning Approaches for Microbiome-Metabolomics Integration with Stable Feature Selection.bioRxiv [Preprint]. 2025 Jun 30:2025.06.21.660858. doi: 10.1101/2025.06.21.660858. bioRxiv. 2025. PMID: 40631202 Free PMC article. Preprint.
-
Crosstalk between gut microbiota and host immune system and its response to traumatic injury.Front Immunol. 2024 Jul 31;15:1413485. doi: 10.3389/fimmu.2024.1413485. eCollection 2024. Front Immunol. 2024. PMID: 39144142 Free PMC article. Review.
-
Semisynthetic simulation for microbiome data analysis.Brief Bioinform. 2024 Nov 22;26(1):bbaf051. doi: 10.1093/bib/bbaf051. Brief Bioinform. 2024. PMID: 39927858 Free PMC article. Review.
-
Advancing Gut Microbiome Research: The Shift from Metagenomics to Multi-Omics and Future Perspectives.J Microbiol Biotechnol. 2025 Mar 26;35:e2412001. doi: 10.4014/jmb.2412.12001. J Microbiol Biotechnol. 2025. PMID: 40223273 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
