Asymmetric and parallel subgenome selection co-shape common carp domestication
- PMID: 38166816
- PMCID: PMC10762839
- DOI: 10.1186/s12915-023-01806-9
Asymmetric and parallel subgenome selection co-shape common carp domestication
Abstract
Background: The common carp (Cyprinus carpio) might best represent the domesticated allopolyploid animals. Although subgenome divergence which is well-known to be a key to allopolyploid domestication has been comprehensively characterized in common carps, the link between genetic architecture underlying agronomic traits and subgenome divergence is unknown in the selective breeding of common carps globally.
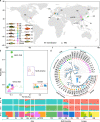
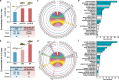
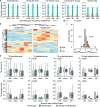
Results: We utilized a comprehensive SNP dataset in 13 representative common carp strains worldwide to detect genome-wide genetic variations associated with scale reduction, vibrant skin color, and high growth rate in common carp domestication. We identified numerous novel candidate genes underlie the three agronomically most desirable traits in domesticated common carps, providing potential molecular targets for future genetic improvement in the selective breeding of common carps. We found that independently selective breeding of the same agronomic trait (e.g., fast growing) in common carp domestication could result from completely different genetic variations, indicating the potential advantage of allopolyploid in domestication. We observed that candidate genes associated with scale reduction, vibrant skin color, and/or high growth rate are repeatedly enriched in the immune system, suggesting that domestication of common carps was often accompanied by the disease resistance improvement.
Conclusions: In common carp domestication, asymmetric subgenome selection is prevalent, while parallel subgenome selection occurs in selective breeding of common carps. This observation is not due to asymmetric gene retention/loss between subgenomes but might be better explained by reduced pleiotropy through transposable element-mediated expression divergence between ohnologs. Our results demonstrate that domestication benefits from polyploidy not only in plants but also in animals.
Keywords: Allopolyploid; High growth rate; Scale reduction; Selection sweep; Vibrant skin color.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Intercross population study reveals that co-mutation of mitfa genes in two subgenomes induces red skin color in common carp ( Cyprinus carpio wuyuanensis).Zool Res. 2023 Mar 18;44(2):276-286. doi: 10.24272/j.issn.2095-8137.2022.388. Zool Res. 2023. PMID: 36785895 Free PMC article.
-
Selection pressures have driven population differentiation of domesticated and wild common carp (Cyprinus carpio L.).Genet Mol Res. 2012 Sep 12;11(3):3222-35. doi: 10.4238/2012.September.12.5. Genet Mol Res. 2012. PMID: 23079816
-
Symmetric expression of ohnologs encoding conserved antiviral responses in tetraploid common carp suggest absence of subgenome dominance after whole genome duplication.Genomics. 2023 Nov;115(6):110723. doi: 10.1016/j.ygeno.2023.110723. Epub 2023 Oct 5. Genomics. 2023. PMID: 37804957
-
Phenotype transition from wild mouflon to domestic sheep.Genet Sel Evol. 2024 Jan 2;56(1):1. doi: 10.1186/s12711-023-00871-6. Genet Sel Evol. 2024. PMID: 38166592 Free PMC article. Review.
-
The role of G protein-coupled receptors and their ligands in animal domestication.Anim Genet. 2024 Dec;55(6):893-906. doi: 10.1111/age.13476. Epub 2024 Sep 26. Anim Genet. 2024. PMID: 39324206 Review.
Cited by
-
Parallel Selection in Domesticated Atlantic Salmon from Divergent Founders Including on Whole-Genome Duplication-derived Homeologous Regions.Genome Biol Evol. 2025 Apr 3;17(4):evaf063. doi: 10.1093/gbe/evaf063. Genome Biol Evol. 2025. PMID: 40247730 Free PMC article.
References
-
- He F, Wang W, Rutter WB, Jordan KW, Ren J, Taagen E, DeWitt N, Sehgal D, Sukumaran S, Dreisigacker S, et al. Genomic variants affecting homoeologous gene expression dosage contribute to agronomic trait variation in allopolyploid wheat. Nat Commun. 2022;13:826. doi: 10.1038/s41467-022-28453-y. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

