Transcriptomic analysis identifies diagnostic genes in polycystic ovary syndrome and periodontitis
- PMID: 38167332
- PMCID: PMC10762819
- DOI: 10.1186/s40001-023-01499-4
Transcriptomic analysis identifies diagnostic genes in polycystic ovary syndrome and periodontitis
Abstract
Purpose: To investigate underlying co-mechanisms of PCOS and periodontitis through transcriptomic approach.
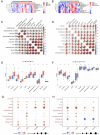
Methods: PCOS and periodontitis gene expression data were downloaded from the GEO database to identify differentially expressed genes. GO and KEGG pathway enrichment analysis and random forest algorithm were used to screen hub genes. GSEA analyzed the functions of hub genes. Correlations between hub genes and immune infiltration in two diseases were examined, constructing a TF-ceRNA regulatory network. Clinical samples were gathered from PCOS and periodontitis patients and RT-qPCR was performed to verify the connection.
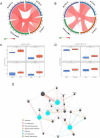
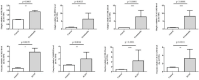
Results: There were 1661 DEGs in PCOS and 701 DEGs in periodontitis. 66 intersected genes were involved and were enriched in immune and inflammation-related biological pathways. 40 common genes were selected from the PPI network. RF algorithm demonstrated that ACSL5, NLRP12, CCRL2, and CEACAM3 were hub genes, and GSEA results revealed their close relationship with antigen processing and presentation, and chemokine signaling pathway. RT-qPCR results confirmed the upregulated gene expression in both PCOS and periodontitis.
Conclusion: The 4 hub genes ACSL5, NLRP12, CCRL2, and CEACAM3 may be diagnostic genes for PCOS and periodontitis. The created ceRNA network could provide a molecular basis for future studies on the association between PCOS and periodontitis.
Keywords: Diagnosis; GSEA; Periodontitis; Polycystic ovary syndrome; Transcriptomic analysis.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
Identification of Hub Genes and Biomarkers between Hyperandrogen and Normoandrogen Polycystic Ovary Syndrome by Bioinformatics Analysis.Comb Chem High Throughput Screen. 2023;26(1):126-134. doi: 10.2174/1386207325666220404101009. Comb Chem High Throughput Screen. 2023. PMID: 35379124
-
Development of machine learning models for diagnostic biomarker identification and immune cell infiltration analysis in PCOS.J Ovarian Res. 2025 Jan 3;18(1):1. doi: 10.1186/s13048-024-01583-1. J Ovarian Res. 2025. PMID: 39754246 Free PMC article.
-
Integrated bioinformatics analysis elucidates granulosa cell whole-transcriptome landscape of PCOS in China.J Ovarian Res. 2023 Aug 3;16(1):154. doi: 10.1186/s13048-023-01223-0. J Ovarian Res. 2023. PMID: 37537636 Free PMC article.
-
Identification of key biomarkers for predicting atherosclerosis progression in polycystic ovary syndrome via bioinformatics analysis and machine learning.Comput Biol Med. 2024 Dec;183:109239. doi: 10.1016/j.compbiomed.2024.109239. Epub 2024 Oct 12. Comput Biol Med. 2024. PMID: 39396400
-
Explore the potential molecular mechanism of polycystic ovarian syndrome by protein-protein interaction network analysis.Taiwan J Obstet Gynecol. 2021 Sep;60(5):807-815. doi: 10.1016/j.tjog.2021.07.005. Taiwan J Obstet Gynecol. 2021. PMID: 34507653 Review.
Cited by
-
Metabolism-related proteins as biomarkers for predicting prognosis in polycystic ovary syndrome.Proteome Sci. 2024 Dec 19;22(1):14. doi: 10.1186/s12953-024-00238-9. Proteome Sci. 2024. PMID: 39702179 Free PMC article.
-
Artificial intelligence in polycystic ovarian syndrome management: past, present, and future.Radiol Med. 2025 Jun 23. doi: 10.1007/s11547-025-02032-9. Online ahead of print. Radiol Med. 2025. PMID: 40549330 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

