A distant global control region is essential for normal expression of anterior HOXA genes during mouse and human craniofacial development
- PMID: 38167838
- PMCID: PMC10762089
- DOI: 10.1038/s41467-023-44506-2
A distant global control region is essential for normal expression of anterior HOXA genes during mouse and human craniofacial development
Abstract
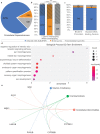
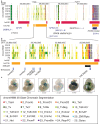
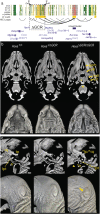
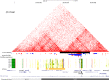
Craniofacial abnormalities account for approximately one third of birth defects. The regulatory programs that build the face require precisely controlled spatiotemporal gene expression, achieved through tissue-specific enhancers. Clusters of coactivated enhancers and their target genes, known as superenhancers, are important in determining cell identity but have been largely unexplored in development. In this study we identified superenhancer regions unique to human embryonic craniofacial tissue. To demonstrate the importance of such regions in craniofacial development and disease, we focused on an ~600 kb noncoding region located between NPVF and NFE2L3. We identified long range interactions with this region in both human and mouse embryonic craniofacial tissue with the anterior portion of the HOXA gene cluster. Mice lacking this superenhancer exhibit perinatal lethality, and present with highly penetrant skull defects and orofacial clefts phenocopying Hoxa2-/- mice. Moreover, we identified two cases of de novo copy number changes of the superenhancer in humans both with severe craniofacial abnormalities. This evidence suggests we have identified a critical noncoding locus control region that specifically regulates anterior HOXA genes and copy number changes are pathogenic in human patients.
© 2024. The Author(s).
Conflict of interest statement
DEM holds stock options in MyOme. DEM is on a scientific advisory board at ONT, is engaged in a research agreement with ONT, and they have paid for him to travel to speak on their behalf.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

