Genome-wide association studies for economically important traits in mink using copy number variation
- PMID: 38167844
- PMCID: PMC10762091
- DOI: 10.1038/s41598-023-50497-3
Genome-wide association studies for economically important traits in mink using copy number variation
Abstract
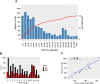
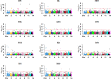
Copy number variations (CNVs) are structural variants consisting of duplications and deletions of DNA segments, which are known to play important roles in the genetics of complex traits in livestock species. However, CNV-based genome-wide association studies (GWAS) have remained unexplored in American mink. Therefore, the purpose of the current study was to investigate the association between CNVs and complex traits in American mink. A CNV-based GWAS was performed with the ParseCNV2 software program using deregressed estimated breeding values of 27 traits as pseudophenotypes, categorized into traits of growth and feed efficiency, reproduction, pelt quality, and Aleutian disease tests. The study identified a total of 10,137 CNVs (6968 duplications and 3169 deletions) using the Affymetrix Mink 70K single nucleotide polymorphism (SNP) array in 2986 American mink. The association analyses identified 250 CNV regions (CNVRs) associated with at least one of the studied traits. These CNVRs overlapped with a total of 320 potential candidate genes, and among them, several genes have been known to be related to the traits such as ARID1B, APPL1, TOX, and GPC5 (growth and feed efficiency traits); GRM1, RNASE10, WNT3, WNT3A, and WNT9B (reproduction traits); MYO10, and LIMS1 (pelt quality traits); and IFNGR2, APEX1, UBE3A, and STX11 (Aleutian disease tests). Overall, the results of the study provide potential candidate genes that may regulate economically important traits and therefore may be used as genetic markers in mink genomic breeding programs.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Genome-wide detection of copy number variation in American mink using whole-genome sequencing.BMC Genomics. 2022 Sep 13;23(1):649. doi: 10.1186/s12864-022-08874-1. BMC Genomics. 2022. PMID: 36096727 Free PMC article.
-
Identification of consensus homozygous regions and their associations with growth and feed efficiency traits in American mink.BMC Genom Data. 2024 Jul 10;25(1):68. doi: 10.1186/s12863-024-01252-8. BMC Genom Data. 2024. PMID: 38982354 Free PMC article.
-
Genome-wide detection of CNVs and their association with performance traits in broilers.BMC Genomics. 2021 May 17;22(1):354. doi: 10.1186/s12864-021-07676-1. BMC Genomics. 2021. PMID: 34001004 Free PMC article.
-
Genome wide copy number variations using Porcine 60K SNP Beadchip in Landlly pigs.Anim Biotechnol. 2023 Nov;34(6):1891-1899. doi: 10.1080/10495398.2022.2056047. Epub 2022 Apr 4. Anim Biotechnol. 2023. PMID: 35369845 Review.
-
Advancements in copy number variation screening in herbivorous livestock genomes and their association with phenotypic traits.Front Vet Sci. 2024 Jan 11;10:1334434. doi: 10.3389/fvets.2023.1334434. eCollection 2023. Front Vet Sci. 2024. PMID: 38274664 Free PMC article. Review.
Cited by
-
Analysis of genomic selection characteristics of local cattle breeds in Gansu.BMC Genomics. 2025 Jul 1;26(1):574. doi: 10.1186/s12864-025-11753-0. BMC Genomics. 2025. PMID: 40597588 Free PMC article.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

