A candidate antibody drug for prevention of malaria
- PMID: 38167935
- PMCID: PMC10803262
- DOI: 10.1038/s41591-023-02659-z
A candidate antibody drug for prevention of malaria
Abstract
Over 75% of malaria-attributable deaths occur in children under the age of 5 years. However, the first malaria vaccine recommended by the World Health Organization (WHO) for pediatric use, RTS,S/AS01 (Mosquirix), has modest efficacy. Complementary strategies, including monoclonal antibodies, will be important in efforts to eradicate malaria. Here we characterize the circulating B cell repertoires of 45 RTS,S/AS01 vaccinees and discover monoclonal antibodies for development as potential therapeutics. We generated >28,000 antibody sequences and tested 481 antibodies for binding activity and 125 antibodies for antimalaria activity in vivo. Through these analyses we identified correlations suggesting that sequences in Plasmodium falciparum circumsporozoite protein, the target antigen in RTS,S/AS01, may induce immunodominant antibody responses that limit more protective, but subdominant, responses. Using binding studies, mouse malaria models, biomanufacturing assessments and protein stability assays, we selected AB-000224 and AB-007088 for advancement as a clinical lead and backup. We engineered the variable domains (Fv) of both antibodies to enable low-cost manufacturing at scale for distribution to pediatric populations, in alignment with WHO's preferred product guidelines. The engineered clone with the optimal manufacturing and drug property profile, MAM01, was advanced into clinical development.
© 2024. The Author(s).
Conflict of interest statement
W.H.R. owns equity in, serves as a consultant to and is a member of the Board of Directors of Atreca, Inc. K.L.W., S.G., K.S.W. and S.M.L. own equity in, and are employed by, Atreca, Inc. D.E.E. and M.W. own equity in, and serve as consultants to, Atreca, Inc. G.C. and D.K. own equity in Atreca, Inc. C.S., P.S., S.Z.J. and R.R.K. are employed by Just – Evotec Biologics. E.J. owns equity in, and is employed by, GSK. U.W.-R. is employed by BioNTech. J.G. owns equity in, is employed by and is a member of the Board of Directors of Centivax, Inc. Y.C.T. owns equity in Atreca, Inc. and is employed by Nuevocor Pte. Ltd. K.L.W., S.M.L., R.R.K. and D.E.E. are coinventors on patent applications filed by Atreca, Inc. that include antibodies to CSP. The remaining authors declare no competing interests.
Figures



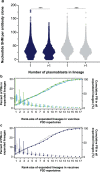
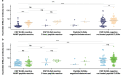


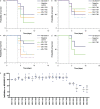
Comment in
-
Integrating vaccines and monoclonal antibodies into malaria prevention.Nat Med. 2024 Jan;30(1):37-38. doi: 10.1038/s41591-023-02745-2. Nat Med. 2024. PMID: 38200257 No abstract available.
References
-
- World Health Organization. World malaria report 2022. https://www.who.int/teams/global-malaria-programme/reports/world-malaria... (2022).
-
- World Health Organization. Global technical strategy for malaria 2016-2030, 2021 update. www.who.int/publications/i/item/9789240031357 (2021).
-
- World Health Organization. WHO recommends R21/Matrix-M vaccine for malaria prevention in updated advice on immunization. www.who.int/news/item/02-10-2023-who-recommends-r21-matrix-m-vaccine-for... (2023).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

