Gut microbiota facilitate chronic spontaneous urticaria
- PMID: 38168034
- PMCID: PMC10762022
- DOI: 10.1038/s41467-023-44373-x
Gut microbiota facilitate chronic spontaneous urticaria
Abstract
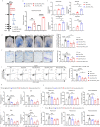
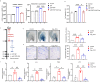
Chronic spontaneous urticaria (CSU) comes with gut dysbiosis, but its relevance remains elusive. Here we use metagenomics sequencing and short-chain fatty acids metabolomics and assess the effects of human CSU fecal microbial transplantation, Klebsiella pneumoniae, Roseburia hominis, and metabolites in vivo. CSU gut microbiota displays low diversity and short-chain fatty acids production, but high gut Klebsiella pneumoniae levels, negatively correlates with blood short-chain fatty acids levels and links to high disease activity. Blood lipopolysaccharide levels are elevated, link to rapid disease relapse, and high gut levels of conditional pathogenic bacteria. CSU microbiome transfer and Klebsiella pneumoniae transplantation facilitate IgE-mediated mast cell(MC)-driven skin inflammatory responses and increase intestinal permeability and blood lipopolysaccharide accumulation in recipient mice. Transplantation of Roseburia hominis and caproate administration protect recipient mice from MC-driven skin inflammation. Here, we show gut microbiome alterations, in CSU, may reduce short-chain fatty acids and increase lipopolysaccharide levels, respectively, and facilitate MC-driven skin inflammation.
© 2024. The Author(s).
Conflict of interest statement
M.Ma is or recently was a speaker and/or advisor for and/or has received research funding from Astria, Allakos, Alnylam, Amgen, Aralez, ArgenX, AstraZeneca, BioCryst, Blueprint, Celldex, Centogene, CSL Behring, Dyax, FAES, Genentech, GIInnovation, GSK, Innate Pharma, Kalvista, Kyowa Kirin, Leo Pharma, Lilly, Menarini, Moxie, Novartis, Pfizer, Pharming, Pharvaris, Roche, Sanofi/Regeneron, Shire/Takeda, Third Harmonic Bio, UCB, and Uriach. All other authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- 81974476/National Natural Science Foundation of China (National Science Foundation of China)
- 82173424/National Natural Science Foundation of China (National Science Foundation of China)
- 82073458/National Natural Science Foundation of China (National Science Foundation of China)
- 81830096/National Natural Science Foundation of China (National Science Foundation of China)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

