The antibiotic resistance reservoir of the lung microbiome expands with age in a population of critically ill patients
- PMID: 38168095
- PMCID: PMC10762195
- DOI: 10.1038/s41467-023-44353-1
The antibiotic resistance reservoir of the lung microbiome expands with age in a population of critically ill patients
Abstract
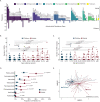
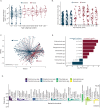
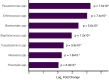
Antimicrobial resistant lower respiratory tract infections are an increasing public health threat and an important cause of global mortality. The lung microbiome can influence susceptibility of respiratory tract infections and represents an important reservoir for exchange of antimicrobial resistance genes. Studies of the gut microbiome have found an association between age and increasing antimicrobial resistance gene burden, however, corollary studies in the lung microbiome remain absent. We performed an observational study of children and adults with acute respiratory failure admitted to the intensive care unit. From tracheal aspirate RNA sequencing data, we evaluated age-related differences in detectable antimicrobial resistance gene expression in the lung microbiome. Using a multivariable logistic regression model, we find that detection of antimicrobial resistance gene expression was significantly higher in adults compared with children after adjusting for demographic and clinical characteristics. This association remained significant after additionally adjusting for lung bacterial microbiome characteristics, and when modeling age as a continuous variable. The proportion of adults expressing beta-lactam, aminoglycoside, and tetracycline antimicrobial resistance genes was higher compared to children. Together, these findings shape our understanding of the lung resistome in critically ill patients across the lifespan, which may have implications for clinical management and global public health.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interest.
Figures


Update of
-
The antibiotic resistance reservoir of the lung microbiome expands with age.Res Sq [Preprint]. 2023 Sep 18:rs.3.rs-3283415. doi: 10.21203/rs.3.rs-3283415/v1. Res Sq. 2023. Update in: Nat Commun. 2024 Jan 2;15(1):92. doi: 10.1038/s41467-023-44353-1. PMID: 37790384 Free PMC article. Updated. Preprint.

