This is a preprint.
Optimizing representations for integrative structural modeling using Bayesian model selection
- PMID: 38168172
- PMCID: PMC10760022
- DOI: 10.1101/2023.12.12.571227
Optimizing representations for integrative structural modeling using Bayesian model selection
Update in
-
Optimizing representations for integrative structural modeling using Bayesian model selection.Bioinformatics. 2024 Mar 4;40(3):btae106. doi: 10.1093/bioinformatics/btae106. Bioinformatics. 2024. PMID: 38391029 Free PMC article.
Abstract
Motivation: Integrative structural modeling combines data from experiments, physical principles, statistics of previous structures, and prior models to obtain structures of macromolecular assemblies that are challenging to characterize experimentally. The choice of model representation is a key decision in integrative modeling, as it dictates the accuracy of scoring, efficiency of sampling, and resolution of analysis. But currently, the choice is usually made ad hoc, manually.
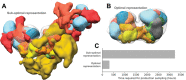
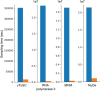
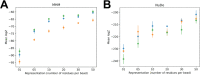
Results: Here, we report NestOR (Nested Sampling for Optimizing Representation), a fully automated, statistically rigorous method based on Bayesian model selection to identify the optimal coarse-grained representation for a given integrative modeling setup. Given an integrative modeling setup, it determines the optimal representations from given candidate representations based on their model evidence and sampling efficiency. The performance of NestOR was evaluated on a benchmark of four macromolecular assemblies.
Availability: NestOR is implemented in the Integrative Modeling Platform (https://integrativemodeling.org) and is available at https://github.com/isblab/nestor.
Keywords: Bayes factors; Bayesian model selection; coarse-grained representation; integrative modeling; macromolecular assemblies; model evidence; nested sampling.
Conflict of interest statement
Conflict of Interest None declared.
Figures

Similar articles
-
Optimizing representations for integrative structural modeling using Bayesian model selection.Bioinformatics. 2024 Mar 4;40(3):btae106. doi: 10.1093/bioinformatics/btae106. Bioinformatics. 2024. PMID: 38391029 Free PMC article.
-
Optimizing model representation for integrative structure determination of macromolecular assemblies.Proc Natl Acad Sci U S A. 2019 Jan 8;116(2):540-545. doi: 10.1073/pnas.1814649116. Epub 2018 Dec 26. Proc Natl Acad Sci U S A. 2019. PMID: 30587581 Free PMC article.
-
Modeling Biological Complexes Using Integrative Modeling Platform.Methods Mol Biol. 2019;2022:353-377. doi: 10.1007/978-1-4939-9608-7_15. Methods Mol Biol. 2019. PMID: 31396911
-
From integrative structural biology to cell biology.J Biol Chem. 2021 Jan-Jun;296:100743. doi: 10.1016/j.jbc.2021.100743. Epub 2021 May 4. J Biol Chem. 2021. PMID: 33957123 Free PMC article. Review.
-
Bayesian methods in integrative structure modeling.Biol Chem. 2023 Jul 31;404(8-9):741-754. doi: 10.1515/hsz-2023-0145. Print 2023 Jul 26. Biol Chem. 2023. PMID: 37505205 Review.
References
-
- Alber F, Dokudovskaya S, Veenhoff LM et al. Determining the architectures of macromolecular assemblies. Nature 2007;450:683–94. - PubMed
-
- Ashton G, Bernstein N, Buchner J et al. Nested sampling for physical scientists. Nat Rev Methods Primers 2022;2:1–22.
-
- Bonomi M, Heller GT, Camilloni C et al. Principles of protein structural ensemble determination. Current Opinion in Structural Biology 2017;42:106–16. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
