This is a preprint.
Epigenetic disruption of the RARγ complex impairs its function to bookmark AR enhancer interactions required for enzalutamide sensitivity in prostate cancer
- PMID: 38168185
- PMCID: PMC10760102
- DOI: 10.1101/2023.12.15.571947
Epigenetic disruption of the RARγ complex impairs its function to bookmark AR enhancer interactions required for enzalutamide sensitivity in prostate cancer
Abstract
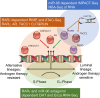
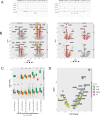
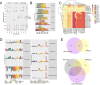
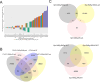
The current study in prostate cancer (PCa) focused on the genomic mechanisms at the cross-roads of pro-differentiation signals and the emergence of lineage plasticity. We explored an understudied cistromic mechanism involving RARγ's ability to govern AR cistrome-transcriptome relationships, including those associated with more aggressive PCa features. The RARγ complex in PCa cell models was enriched for canonical cofactors, as well as proteins involved in RNA processing and bookmarking. Identifying the repertoire of miR-96 bound and regulated gene targets, including those recognition elements marked by m6A, revealed their significant enrichment in the RARγ complex. RARγ significantly enhanced the AR cistrome, particularly in active enhancers and super-enhancers, and overlapped with the binding of bookmarking factors. Furthermore, RARγ expression led to nucleosome-free chromatin enriched with H3K27ac, and significantly enhanced the AR cistrome in G2/M cells. RARγ functions also antagonized the transcriptional actions of the lineage master regulator ONECUT2. Similarly, gene programs regulated by either miR-96 or antagonized by RARγ were enriched in alternative lineages and more aggressive PCa phenotypes. Together these findings reveal an under-investigated role for RARγ, modulated by miR-96, to bookmark enhancer sites during mitosis. These sites are required by the AR to promote transcriptional competence, and emphasize luminal differentiation, while antagonizing ONECUT2.
Conflict of interest statement
Competing interests The authors certify that they has NO affiliations with or involvement in any organization or entity with any financial interest (such as honoraria; educational grants; participation in speakers’ bureaus; membership, employment, consultancies, stock ownership, or other equity interest; and expert testimony or patent-licensing arrangements), or non-financial interest (such as personal or professional relationships, affiliations, knowledge or beliefs) in the subject matter or materials discussed in this manuscript.
Figures



References
-
- Ellis L. (2017) Understanding cancer lineage plasticity: reversing therapeutic resistance in metastatic prostate cancer. Pharmacogenomics, 18, 597–600. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
