This is a preprint.
Machine learning models for blood pressure phenotypes combining multiple polygenic risk scores
- PMID: 38168328
- PMCID: PMC10760279
- DOI: 10.1101/2023.12.13.23299909
Machine learning models for blood pressure phenotypes combining multiple polygenic risk scores
Update in
-
Machine learning models for predicting blood pressure phenotypes by combining multiple polygenic risk scores.Sci Rep. 2024 May 30;14(1):12436. doi: 10.1038/s41598-024-62945-9. Sci Rep. 2024. PMID: 38816422 Free PMC article.
Abstract
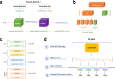
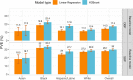
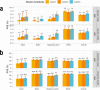
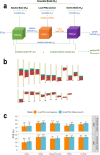
We construct non-linear machine learning (ML) prediction models for systolic and diastolic blood pressure (SBP, DBP) using demographic and clinical variables and polygenic risk scores (PRSs). We developed a two-model ensemble, consisting of a baseline model, where prediction is based on demographic and clinical variables only, and a genetic model, where we also include PRSs. We evaluate the use of a linear versus a non-linear model at both the baseline and the genetic model levels and assess the improvement in performance when incorporating multiple PRSs. We report the ensemble model's performance as percentage variance explained (PVE) on a held-out test dataset. A non-linear baseline model improved the PVEs from 28.1% to 30.1% (SBP) and 14.3% to 17.4% (DBP) compared with a linear baseline model. Including seven PRSs in the genetic model computed based on the largest available GWAS of SBP/DBP improved the genetic model PVE from 4.8% to 5.1% (SBP) and 4.7% to 5% (DBP) compared to using a single PRS. Adding additional 14 PRSs computed based on two independent GWASs further increased the genetic model PVE to 6.3% (SBP) and 5.7% (DBP). PVE differed across self-reported race/ethnicity groups, with primarily all non-White groups benefitting from the inclusion of additional PRSs.
Conflict of interest statement
Conflict of interests B Psaty serves on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson. G Lyons is currently an employee of Alexion Pharmaceuticals, however, her contributions to the present manuscript were performed as part of her previous affiliation at the Harvard T.H. Chan School of Public Health and this work is not related to her current occupation and affiliation. M Moll has received grant funding from Bayer and consulting fees from TriNetX, 2ndMD, TheaHealth, Sitka, Verona Pharma, and Axon Advisors.
Figures
References
-
- Torkamani A., Wineinger N.E., and Topol E.J., The personal and clinical utility of polygenic risk scores. Nat Rev Genet, 2018. 19(9): p. 581–590. - PubMed
-
- Tibshirani R., Regression Shrinkage and Selection via the Lasso. Journal of the Royal Statistical Society. Series B (Methodological), 1996. 58(1): p. 267–288.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
