This is a preprint.
Biased cell adhesion organizes a circuit for visual motion integration
- PMID: 38168373
- PMCID: PMC10760042
- DOI: 10.1101/2023.12.11.571076
Biased cell adhesion organizes a circuit for visual motion integration
Update in
-
Biased cell adhesion organizes the Drosophila visual motion integration circuit.Dev Cell. 2025 Mar 10;60(5):762-779.e7. doi: 10.1016/j.devcel.2024.10.019. Epub 2024 Nov 15. Dev Cell. 2025. PMID: 39549704
Abstract
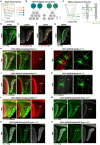
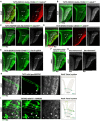
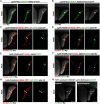
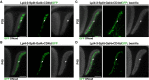
Layer specific computations in the brain rely on neuronal processes establishing synaptic connections with specific partners in distinct laminae. In the Drosophila lobula plate neuropile, the axons of the four subtypes of T4 and T5 visual motion direction-selective neurons segregate into four layers, based on their directional preference, and form synapses with distinct subsets of postsynaptic neurons. Four bi-stratified inhibitory lobula plate intrinsic cells exhibit a consistent synaptic pattern, receiving excitatory T4/T5 inputs in one layer, and conveying inhibitory signals to an adjacent layer. This layered arrangement establishes motion opponency. Here, we identify layer-specific expression of different receptor-ligand pairs belonging to the Beat and Side families of Cell Adhesion Molecules (CAMs) between T4/T5 neurons and their postsynaptic partners. Genetic analysis reveals that Beat/Side mediated interactions are required to restrict T4/T5 axonal innervation to a single layer. We propose that Beat/Side contribute to synaptic specificity by biasing adhesion between synaptic partners before synaptogenesis.
Figures



References
-
- Baier H. (2013). Synaptic laminae in the visual system: molecular mechanisms forming layers of perception. Annual review of cell and developmental biology 29, 385–416. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
