This is a preprint.
A protective and broadly binding antibody class engages the influenza virus hemagglutinin head at its stem interface
- PMID: 38168412
- PMCID: PMC10760138
- DOI: 10.1101/2023.12.13.571543
A protective and broadly binding antibody class engages the influenza virus hemagglutinin head at its stem interface
Update in
-
A protective and broadly binding antibody class engages the influenza virus hemagglutinin head at its stem interface.mBio. 2025 Jun 11;16(6):e0089225. doi: 10.1128/mbio.00892-25. Epub 2025 May 20. mBio. 2025. PMID: 40391889 Free PMC article.
Abstract
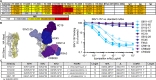
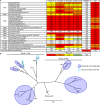
Influenza infection and vaccination impart strain-specific immunity that protects against neither seasonal antigenic variants nor the next pandemic. However, antibodies directed to conserved sites can confer broad protection. Here we identify and characterize a class of human antibodies that engage a previously undescribed, conserved epitope on the influenza hemagglutinin (HA) protein. Prototype antibody S8V1-157 binds at the normally occluded interface between the HA head and stem. Antibodies to this HA head-stem interface epitope are non-neutralizing in vitro but protect against lethal influenza infection in mice. Antibody isotypes that direct clearance of infected cells enhance this protection. Head-stem interface antibodies bind to most influenza A serotypes and seasonal human variants, and are present at low frequencies in the memory B cell populations of multiple human donors. Vaccines designed to elicit these antibodies might contribute to "universal" influenza immunity.
Figures




References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
