This is a preprint.
A germline chimeric KANK1-DMRT1 transcript derived from a complex structural variant is associated with a congenital heart defect segregating across five generations
- PMID: 38168413
- PMCID: PMC10760254
- DOI: 10.21203/rs.3.rs-3740005/v1
A germline chimeric KANK1-DMRT1 transcript derived from a complex structural variant is associated with a congenital heart defect segregating across five generations
Update in
-
A germline chimeric KANK1-DMRT1 transcript derived from a complex structural variant is associated with a congenital heart defect segregating across five generations.Chromosome Res. 2024 Mar 19;32(2):6. doi: 10.1007/s10577-024-09750-2. Chromosome Res. 2024. PMID: 38504027
Abstract
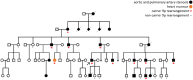
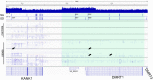
Structural variants (SVs) pose a challenge to detect and interpret, but their study provides novel biological insights and molecular diagnosis underlying rare diseases. The aim of this study was to resolve a 9p24 rearrangement segregating in a family through five generations with a congenital heart defect (congenital pulmonary and aortic valvular stenosis, and pulmonary artery stenosis), by applying a combined genomic analysis. The analysis involved multiple techniques, including karyotype, chromosomal microarray analysis (CMA), FISH, whole-genome sequencing (WGS), RNA-seq and optical genome mapping (OGM). A complex 9p24 SV was hinted at by CMA results, showing three interspersed duplicated segments. Combined WGS and OGM analyses revealed that the 9p24 duplications constitute a complex SV, on which a set of breakpoints match the boundaries of the CMA duplicated sequences. The proposed structure for this complex rearrangement implies three duplications associated with an inversion of ~ 2Mb region on chromosome 9 with a SINE element insertion at the more distal breakpoint. Interestingly, this hypothesized genomic structure of rearrangement forms a chimeric transcript of the KANK1/DMRT1 loci, which was confirmed by RNA-seq on blood from 9p24 rearrangement carriers. Altogether with breakpoint amplification and FISH analysis, this combined approach allowed a deep characterization of this complex rearrangement. Although the genotype-phenotype correlation remains elusive from the molecular mechanism point of view, this study identified a large genomic rearrangement at 9p segregating with a familial congenital clinical trait, revealing a genetic biomarker that was successfully applied for embryo selection, changing the reproductive perspective of affected individuals.
Keywords: KANK1; congenital pulmonary and aortic valvular stenosis; germline quimeric transcripts; pulmonary artery stenosis; structural variation.
Conflict of interest statement
Conflicts of interest/Competing interests The authors have no relevant financial or non-financial interests to disclose.
Figures



References
-
- Boone Philip M., Yuan Bo, Campbell Ian M., Scull Jennifer C., Withers Marjorie A., Baggett Brett C., Beck Christine R., et al. 2014. “The Alu-Rich Genomic Architecture of SPAST Predisposes to Diverse and Functionally Distinct Disease-Associated CNV Alleles.” American Journal of Human Genetics 95 (2): 143–61. 10.1016/j.ajhg.2014.06.014. - DOI - PMC - PubMed
-
- Botos Marius Alexandru, Arora Prateek, Chouvardas Panagiotis, and Mercader Nadia. 2023. “Transcriptomic Data Meta-Analysis Reveals Common and Injury Model Specific Gene Expression Changes in the Regenerating Zebrafish Heart.” Scientific Reports 13 (April): 5418. 10.1038/s41598-023-32272-6. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

