This is a preprint.
Predicting the First Steps of Evolution in Randomly Assembled Communities
- PMID: 38168431
- PMCID: PMC10760118
- DOI: 10.1101/2023.12.15.571925
Predicting the First Steps of Evolution in Randomly Assembled Communities
Update in
-
Predicting the first steps of evolution in randomly assembled communities.Nat Commun. 2024 Oct 1;15(1):8495. doi: 10.1038/s41467-024-52467-3. Nat Commun. 2024. PMID: 39353888 Free PMC article.
Abstract
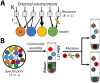
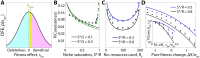
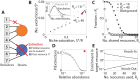
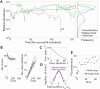
Microbial communities can self-assemble into highly diverse states with predictable statistical properties. However, these initial states can be disrupted by rapid evolution of the resident strains. When a new mutation arises, it competes for resources with its parent strain and with the other species in the community. This interplay between ecology and evolution is difficult to capture with existing community assembly theory. Here, we introduce a mathematical framework for predicting the first steps of evolution in large randomly assembled communities that compete for substitutable resources. We show how the fitness effects of new mutations and the probability that they coexist with their parent depends on the size of the community, the saturation of its niches, and the metabolic overlap between its members. We find that successful mutations are often able to coexist with their parent strains, even in saturated communities with low niche availability. At the same time, these invading mutants often cause extinctions of metabolically distant species. Our results suggest that even small amounts of evolution can produce distinct genetic signatures in natural microbial communities.
Conflict of interest statement
Competing interests: None declared.
Figures

Similar articles
-
Predicting the first steps of evolution in randomly assembled communities.Nat Commun. 2024 Oct 1;15(1):8495. doi: 10.1038/s41467-024-52467-3. Nat Commun. 2024. PMID: 39353888 Free PMC article.
-
The Global Genome Question: Microbes as the Key to Understanding Evolution and Ecology: This report is based on a colloquium, “The Global Genome Question: Microbes as the Key to Understanding Evolution and Ecology,” sponsored by the American Academy of Microbiology and held October 11-13, 2002, in Longboat Key, Florida.Washington (DC): American Society for Microbiology; 2004. Washington (DC): American Society for Microbiology; 2004. PMID: 33119236 Free Books & Documents. Review.
-
Metabolic Overlap in Environmentally Diverse Microbial Communities.Front Genet. 2019 Oct 17;10:989. doi: 10.3389/fgene.2019.00989. eCollection 2019. Front Genet. 2019. PMID: 31681424 Free PMC article.
-
A theory of ecological invasions and its implications for eco-evolutionary dynamics.bioRxiv [Preprint]. 2025 Mar 17:2025.03.14.643284. doi: 10.1101/2025.03.14.643284. bioRxiv. 2025. PMID: 40166292 Free PMC article. Preprint.
-
Can metabolic traits explain animal community assembly and functioning?Biol Rev Camb Philos Soc. 2023 Feb;98(1):1-18. doi: 10.1111/brv.12892. Epub 2022 Aug 25. Biol Rev Camb Philos Soc. 2023. PMID: 36054431 Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
