Candida albicans extracellular vesicles trigger type I IFN signalling via cGAS and STING
- PMID: 38168615
- PMCID: PMC10959075
- DOI: 10.1038/s41564-023-01546-0
Candida albicans extracellular vesicles trigger type I IFN signalling via cGAS and STING
Abstract
The host type I interferon (IFN) pathway is a major signature of inflammation induced by the human fungal pathogen, Candida albicans. However, the molecular mechanism for activating this pathway in the host defence against C. albicans remains unknown. Here we reveal that mice lacking cyclic GMP-AMP synthase (cGAS)-stimulator of IFN genes (STING) pathway components had improved survival following an intravenous challenge by C. albicans. Biofilm-associated C. albicans DNA packaged in extracellular vesicles triggers the cGAS-STING pathway as determined by induction of interferon-stimulated genes, IFNβ production, and phosphorylation of IFN regulatory factor 3 and TANK-binding kinase 1. Extracellular vesicle-induced activation of type I IFNs was independent of the Dectin-1/Card9 pathway and did not require toll-like receptor 9. Single nucleotide polymorphisms in cGAS and STING potently altered inflammatory cytokine production in human monocytes challenged by C. albicans. These studies provide insights into the early innate immune response induced by a clinically significant fungal pathogen.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures





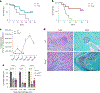

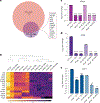

References
-
- Tsay S et al. National burden of candidemia, United States, 2017. Open Forum Infect. Dis 5, S142 (2018).
-
- Pekmezovic M et al. Candida pathogens induce protective mitochondria-associated type I interferon signalling and a damage-driven response in vaginal epithelial cells. Nat. Microbiol 6, 643–657 (2021). - PubMed
-
- Bourgeois C et al. Conventional dendritic cells mount a type I IFN response against Candida spp. Requiring novel phagosomal TLR7-mediated IFN-β signaling. J. Immunol 186, 3104–3112 (2011). - PubMed
MeSH terms
Substances
Grants and funding
- K08 AI141755/AI/NIAID NIH HHS/United States
- R37 AI116550/AI/NIAID NIH HHS/United States
- R01 AI136529/AI/NIAID NIH HHS/United States
- R01 AI150181/AI/NIAID NIH HHS/United States
- R21 AI152499/AI/NIAID NIH HHS/United States
- R01 AI145939/AI/NIAID NIH HHS/United States
- R01 AI153405/AI/NIAID NIH HHS/United States
- R01 AI167993/AI/NIAID NIH HHS/United States
- R01 AI073289/AI/NIAID NIH HHS/United States
- R21 AI156104/AI/NIAID NIH HHS/United States
- R21 AI109303/AI/NIAID NIH HHS/United States
- R21 AI159583/AI/NIAID NIH HHS/United States
- R01 AI092084/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

