Probing the chemical 'reactome' with high-throughput experimentation data
- PMID: 38168924
- PMCID: PMC10997498
- DOI: 10.1038/s41557-023-01393-w
Probing the chemical 'reactome' with high-throughput experimentation data
Abstract
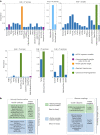
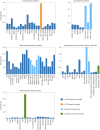
High-throughput experimentation (HTE) has the potential to improve our understanding of organic chemistry by systematically interrogating reactivity across diverse chemical spaces. Notable bottlenecks include few publicly available large-scale datasets and the need for facile interpretation of these data's hidden chemical insights. Here we report the development of a high-throughput experimentation analyser, a robust and statistically rigorous framework, which is applicable to any HTE dataset regardless of size, scope or target reaction outcome, which yields interpretable correlations between starting material(s), reagents and outcomes. We improve the HTE data landscape with the disclosure of 39,000+ previously proprietary HTE reactions that cover a breadth of chemistry, including cross-coupling reactions and chiral salt resolutions. The high-throughput experimentation analyser was validated on cross-coupling and hydrogenation datasets, showcasing the elucidation of statistically significant hidden relationships between reaction components and outcomes, as well as highlighting areas of dataset bias and the specific reaction spaces that necessitate further investigation.
© 2024. The Author(s).
Conflict of interest statement
A.A.L. is a co-founder and owns equity in PostEra Inc and Byterat Ltd. S.B., L.B., X.H., R.M.H., J.L.K., J.M., N.W.S., J.W.T. and Q.Y. are employed by Pfizer Inc. E.K.S. declares no competing interests.
Figures




References
-
- Mennen SM, et al. The evolution of high-throughput experimentation in pharmaceutical development and perspectives on the future. Org. Process Res. Dev. 2019;23:1213–1242. doi: 10.1021/acs.oprd.9b00140. - DOI
LinkOut - more resources
Full Text Sources

