High-quality metagenome assembly from long accurate reads with metaMDBG
- PMID: 38168989
- PMCID: PMC11392814
- DOI: 10.1038/s41587-023-01983-6
High-quality metagenome assembly from long accurate reads with metaMDBG
Abstract
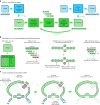
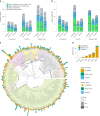
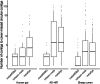
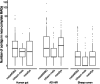
We introduce metaMDBG, a metagenomics assembler for PacBio HiFi reads. MetaMDBG combines a de Bruijn graph assembly in a minimizer space with an iterative assembly over sequences of minimizers to address variations in genome coverage depth and an abundance-based filtering strategy to simplify strain complexity. For complex communities, we obtained up to twice as many high-quality circularized prokaryotic metagenome-assembled genomes as existing methods and had better recovery of viruses and plasmids.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Update of
-
Efficient High-Quality Metagenome Assembly from Long Accurate Reads using Minimizer-space de Bruijn Graphs.bioRxiv [Preprint]. 2023 Jul 8:2023.07.07.548136. doi: 10.1101/2023.07.07.548136. bioRxiv. 2023. Update in: Nat Biotechnol. 2024 Sep;42(9):1378-1383. doi: 10.1038/s41587-023-01983-6. PMID: 37786716 Free PMC article. Updated. Preprint.
References
MeSH terms
Grants and funding
- NE/T013230/1/RCUK | Natural Environment Research Council (NERC)
- MR/S037195/1/RCUK | Medical Research Council (MRC)
- BB/CSP1720/1/RCUK | Biotechnology and Biological Sciences Research Council (BBSRC)
- BS/E/T/000PR9818/RCUK | Biotechnology and Biological Sciences Research Council (BBSRC)
- BBS/E/T/000PR9817/RCUK | Biotechnology and Biological Sciences Research Council (BBSRC)
LinkOut - more resources
Full Text Sources

