Polymyxins with Potent Antibacterial Activity against Colistin-Resistant Pathogens: Fine-Tuning Hydrophobicity with Unnatural Amino Acids
- PMID: 38169430
- PMCID: PMC10824244
- DOI: 10.1021/acs.jmedchem.3c01908
Polymyxins with Potent Antibacterial Activity against Colistin-Resistant Pathogens: Fine-Tuning Hydrophobicity with Unnatural Amino Acids
Abstract
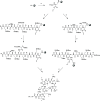
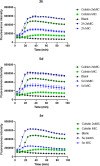
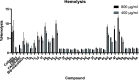
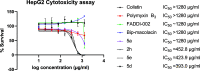
In view of the increased prevalence of antimicrobial resistance among human pathogens, antibiotics against multidrug-resistant (MDR) bacteria are in urgent demand. In particular, the rapidly emerging resistance to last-resort antibiotic colistin, used for severe Gram-negative MDR infections, is critical. Here, a series of polymyxins containing unnatural amino acids were explored, and some analogues exhibited excellent antibacterial activity against Escherichia coli, Klebsiella pneumoniae, Acinetobacter baumannii, and Pseudomonas aeruginosa. Hydrophobicity of the compounds within this series (as measured by retention in reversed-phase analytical HPLC) exhibited a discernible correlation with their antimicrobial activity. This trend was particularly pronounced for colistin-resistant pathogens. The most active compounds demonstrated competitive activity against a panel of Gram-negative pathogens, while exhibiting low in vitro cytotoxicity. Importantly, most of these hits also retained (or even had increased) potency against colistin-susceptible strains. These findings infer that fine-tuning hydrophobicity may enable the design of polymyxin analogues with favorable activity profiles.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

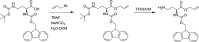

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

