VEGFA/NRP-1/GAPVD1 axis promotes progression and cancer stemness of triple-negative breast cancer by enhancing tumor cell-macrophage crosstalk
- PMID: 38169627
- PMCID: PMC10758102
- DOI: 10.7150/ijbs.86085
VEGFA/NRP-1/GAPVD1 axis promotes progression and cancer stemness of triple-negative breast cancer by enhancing tumor cell-macrophage crosstalk
Abstract
Triple-negative breast cancer (TNBC) has long been considered a major clinical challenge due to its aggressive behavior and poor prognosis. Cancer stem cells (CSCs) are known as the main cells responsible for tumor origination, progression, recurrence and metastasis. Here, we report that M2-type tumor-associated macrophages (TAMs) contribute to cancer stemness in TNBC cells via the secretion of VEGFA. Reciprocally, elevated VEGFA expression by TAM-educated TNBC cells acts as a regulator of macrophage polarization, therefore constitute a feed-back loop between TNBC cells and TAMs. Mechanistically, VEGFA facilitates the CSC phenotype via the NRP-1 receptor and downstream GAPVD1/Wnt/β-catenin signaling pathway in TNBC cells. Our study underscores the crosstalk between TNBC cells and TAMs mediated by VEGFA and further clarifies the role and underlying mechanisms of the VEGFA/NRP-1/GAPVD1 axis in regulating cancer stemness. We also document an immunosuppressive function of VEGFA in the tumor microenvironment (TME). Therefore, the present study indicates crosstalk between TNBC cells and TAMs induced by VEGFA and provides a potential implication for the combination of immunotherapy and VEGFA-targeted agents in TNBC therapy.
Keywords: Triple-negative breast cancer; VEGFA/NRP-1/GAPVD1 axis; cancer stem cell; tumor-associated macrophage..
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures

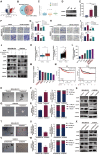
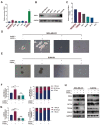
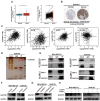



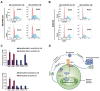
References
-
- Miller KD, Fidler-Benaoudia M, Keegan TH. et al. Cancer statistics for adolescents and young adults, 2020. CA Cancer J Clin. 2020;70(6):443–459. - PubMed
-
- Boyle P. Triple-negative breast cancer: epidemiological considerations and recommendations. Ann Oncol. 2012;23(Suppl 6):vi7–12. - PubMed
-
- Hill BS, Sarnella A, D'Avino G. et al. Recruitment of stromal cells into tumour microenvironment promote the metastatic spread of breast cancer. Semin Cancer Biol. 2020;60:202–213. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

