Characterization of community-acquired Clostridioides difficile strains in Israel, 2020-2022
- PMID: 38169783
- PMCID: PMC10758451
- DOI: 10.3389/fmicb.2023.1323257
Characterization of community-acquired Clostridioides difficile strains in Israel, 2020-2022
Abstract
Background: The prevalence of community-acquired Clostridioides difficile infection (CA-CDI) has been rising, due to changes in antibiotics prescribing practices, emergence of hypervirulent strains and improved diagnostics. This study explored CA-CDI epidemiology by examining strain diversity and virulence factors of CA-CDI isolates collected across several geographical regions in Israel.
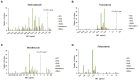
Methods: Stool samples of 126 CA-CDI patients were subjected to PCR and an immunoassay to identify toxin genes and proteins, respectively. Toxin loci PaLoc and PaCdt were detected by whole-genome sequencing (WGS). Biofilm production was assessed by crystal violet-based assay. Minimum inhibitory concentration was determined using the Etest technique or agar dilution. WGS and multi-locus sequence typing (MLST) were used to classify strains and investigate genetic diversity.
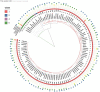
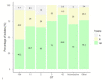
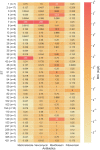
Results: Sequence types (ST) 2 (17, 13.5%), ST42 (13, 10.3%), ST104 (10, 8%) and ST11 (9, 7.1%) were the most common. All (117, 92.8%) but ST11 belonged to Clade 1. No associations were found between ST and gender, geographic area or antibiotic susceptibility. Although all strains harbored toxins genes, 34 (27%) produced toxin A only, and 54 (42.9%) strains produced toxin B only; 38 (30.2%) produced both toxins. Most isolates were biofilm-producers (118, 93.6%), primarily weak producers (83/118, 70.3%). ST was significantly associated with both biofilm and toxin production.
Conclusion: C. difficile isolates in Israel community exhibit high ST diversity, with no dominant strain. Other factors may influence the clinical outcomes of CDI such as toxin production, antibiotic resistance and biofilm production. Further studies are needed to better understand the dynamics and influence of these factors on CA-CDI.
Keywords: C. difficile; CDI; MLST; clade; community-acquired C. difficile infection.
Copyright © 2023 Schwartz, Rohana, Azrad, Shor, Rainy, Maor, Nesher, Sagi, Ken-Dror, Kechker and Peretz.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
LinkOut - more resources
Full Text Sources

