SWOffinder: Efficient and versatile search of CRISPR off-targets with bulges by Smith-Waterman alignment
- PMID: 38169993
- PMCID: PMC10758973
- DOI: 10.1016/j.isci.2023.108557
SWOffinder: Efficient and versatile search of CRISPR off-targets with bulges by Smith-Waterman alignment
Abstract
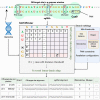
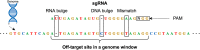
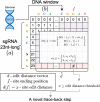
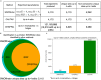
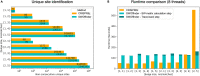
CRISPR/Cas9 technology is revolutionizing the field of gene editing. While this technology enables the targeting of any gene, it may also target unplanned loci, termed off-target sites (OTS), which are a few mismatches, insertions, and deletions from the target. While existing methods for finding OTS up to a given mismatch threshold are efficient, other methods considering insertions and deletions are limited by long runtimes, incomplete OTS lists, and partial support of versatile thresholds. Here, we developed SWOffinder, an efficient method based on Smith-Waterman alignment to find all OTS up to some edit distance. We implemented an original trace-back approach to find OTS under versatile criteria, such as separate limits on the number of insertions, deletions, and mismatches. Compared to state-of-the-art methods, only SWOffinder finds all OTS in the genome in just a few minutes. SWOffinder enables accurate and efficient genomic search of OTS, which will lead to safer gene editing.
Keywords: Algorithms; Bioinformatics; Biological constraints; Techniques in genetics.
© 2023 The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
-
- Jiang F., Doudna J.A. CRISPR–Cas9 structures and mechanisms. Annu. Rev. Biophys. 2017;46:505–529. - PubMed
-
- Pineda M., Lear A., Collins J.P., Kiani S. Safe CRISPR: challenges and possible solutions. Trends Biotechnol. 2019;37:389–401. - PubMed
-
- Backurs A., Indyk P. Proceedings of the forty-seventh annual ACM symposium on Theory of computing. 2015. Edit distance cannot be computed in strongly subquadratic time (unless SETH is false) pp. 51–58.
LinkOut - more resources
Full Text Sources

