Single-cell sequencing dissects the transcriptional identity of activated fibroblasts and identifies novel persistent distal tubular injury patterns in kidney fibrosis
- PMID: 38172172
- PMCID: PMC10764314
- DOI: 10.1038/s41598-023-50195-0
Single-cell sequencing dissects the transcriptional identity of activated fibroblasts and identifies novel persistent distal tubular injury patterns in kidney fibrosis
Abstract
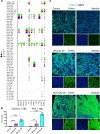
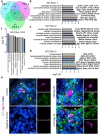
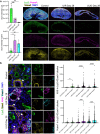
Examining kidney fibrosis is crucial for mechanistic understanding and developing targeted strategies against chronic kidney disease (CKD). Persistent fibroblast activation and tubular epithelial cell (TEC) injury are key CKD contributors. However, cellular and transcriptional landscapes of CKD and specific activated kidney fibroblast clusters remain elusive. Here, we analyzed single cell transcriptomic profiles of two clinically relevant kidney fibrosis models which induced robust kidney parenchymal remodeling. We dissected the molecular and cellular landscapes of kidney stroma and newly identified three distinctive fibroblast clusters with "secretory", "contractile" and "vascular" transcriptional enrichments. Also, both injuries generated failed repair TECs (frTECs) characterized by decline of mature epithelial markers and elevation of stromal and injury markers. Notably, frTECs shared transcriptional identity with distal nephron segments of the embryonic kidney. Moreover, we identified that both models exhibited robust and previously unrecognized distal spatial pattern of TEC injury, outlined by persistent elevation of renal TEC injury markers including Krt8 and Vcam1, while the surviving proximal tubules (PTs) showed restored transcriptional signature. We also found that long-term kidney injuries activated a prominent nephrogenic signature, including Sox4 and Hox gene elevation, which prevailed in the distal tubular segments. Our findings might advance understanding of and targeted intervention in fibrotic kidney disease.
© 2024. The Author(s).
Conflict of interest statement
P.D. is a co-inventor on patents for the use of NGAL as a biomarker of kidney injury. The remaining authors declare no competing interests.
Figures





Update of
-
Single-cell sequencing dissects the transcriptional identity of activated fibroblasts and identifies novel persistent distal tubular injury patterns in kidney fibrosis.Res Sq [Preprint]. 2023 May 17:rs.3.rs-2880248. doi: 10.21203/rs.3.rs-2880248/v1. Res Sq. 2023. Update in: Sci Rep. 2024 Jan 3;14(1):439. doi: 10.1038/s41598-023-50195-0. PMID: 37293022 Free PMC article. Updated. Preprint.
Similar articles
-
Single-cell sequencing dissects the transcriptional identity of activated fibroblasts and identifies novel persistent distal tubular injury patterns in kidney fibrosis.Res Sq [Preprint]. 2023 May 17:rs.3.rs-2880248. doi: 10.21203/rs.3.rs-2880248/v1. Res Sq. 2023. Update in: Sci Rep. 2024 Jan 3;14(1):439. doi: 10.1038/s41598-023-50195-0. PMID: 37293022 Free PMC article. Updated. Preprint.
-
Wnt9a Promotes Renal Fibrosis by Accelerating Cellular Senescence in Tubular Epithelial Cells.J Am Soc Nephrol. 2018 Apr;29(4):1238-1256. doi: 10.1681/ASN.2017050574. Epub 2018 Feb 12. J Am Soc Nephrol. 2018. PMID: 29440280 Free PMC article.
-
Molecular Markers of Tubulointerstitial Fibrosis and Tubular Cell Damage in Patients with Chronic Kidney Disease.PLoS One. 2015 Aug 28;10(8):e0136994. doi: 10.1371/journal.pone.0136994. eCollection 2015. PLoS One. 2015. PMID: 26317775 Free PMC article.
-
[Identification of Biomarkers for Tubular Injury and Interstitial Fibrosis in Chronic Kidney Disease].Yakugaku Zasshi. 2017;137(11):1355-1360. doi: 10.1248/yakushi.17-00150. Yakugaku Zasshi. 2017. PMID: 29093371 Review. Japanese.
-
Kidney fibrosis: from mechanisms to therapeutic medicines.Signal Transduct Target Ther. 2023 Mar 17;8(1):129. doi: 10.1038/s41392-023-01379-7. Signal Transduct Target Ther. 2023. PMID: 36932062 Free PMC article. Review.
Cited by
-
Gucy1α1 specifically marks kidney, heart, lung and liver fibroblasts.Sci Rep. 2024 Nov 26;14(1):29307. doi: 10.1038/s41598-024-80930-0. Sci Rep. 2024. PMID: 39592775 Free PMC article.
-
Insights into Repeated Renal Injury Using RNA-Seq with Two New RPTEC Cell Lines.Int J Mol Sci. 2023 Sep 18;24(18):14228. doi: 10.3390/ijms241814228. Int J Mol Sci. 2023. PMID: 37762531 Free PMC article.
-
Ankrd1 as a potential biomarker for the transition from acute kidney injury to chronic kidney disease.Sci Rep. 2025 Feb 7;15(1):4659. doi: 10.1038/s41598-025-88752-4. Sci Rep. 2025. PMID: 39920300 Free PMC article.
-
Targeting AXL cellular networks in kidney fibrosis.Front Immunol. 2024 Nov 4;15:1446672. doi: 10.3389/fimmu.2024.1446672. eCollection 2024. Front Immunol. 2024. PMID: 39559366 Free PMC article.
-
Targeting the transmembrane cytokine co-receptor neuropilin-1 in distal tubules improves renal injury and fibrosis.Nat Commun. 2024 Jul 8;15(1):5731. doi: 10.1038/s41467-024-50121-6. Nat Commun. 2024. PMID: 38977708 Free PMC article.
References
-
- United States Renal Data System . USRDS Annual Data Report: Epidemiology of Kidney Disease in the United States. National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases; 2021.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

