GIMAP5 deficiency reveals a mammalian ceramide-driven longevity assurance pathway
- PMID: 38172257
- PMCID: PMC11151279
- DOI: 10.1038/s41590-023-01691-y
GIMAP5 deficiency reveals a mammalian ceramide-driven longevity assurance pathway
Erratum in
-
Author Correction: GIMAP5 deficiency reveals a mammalian ceramide-driven longevity assurance pathway.Nat Immunol. 2024 Apr;25(4):717. doi: 10.1038/s41590-024-01779-z. Nat Immunol. 2024. PMID: 38347083 No abstract available.
Abstract
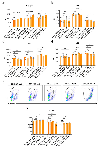
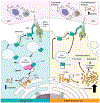
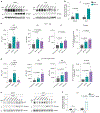
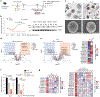
Preserving cells in a functional, non-senescent state is a major goal for extending human healthspans. Model organisms reveal that longevity and senescence are genetically controlled, but how genes control longevity in different mammalian tissues is unknown. Here, we report a new human genetic disease that causes cell senescence, liver and immune dysfunction, and early mortality that results from deficiency of GIMAP5, an evolutionarily conserved GTPase selectively expressed in lymphocytes and endothelial cells. We show that GIMAP5 restricts the pathological accumulation of long-chain ceramides (CERs), thereby regulating longevity. GIMAP5 controls CER abundance by interacting with protein kinase CK2 (CK2), attenuating its ability to activate CER synthases. Inhibition of CK2 and CER synthase rescues GIMAP5-deficient T cells by preventing CER overaccumulation and cell deterioration. Thus, GIMAP5 controls longevity assurance pathways crucial for immune function and healthspan in mammals.
© 2024. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Figures











References
-
- Jiang JC, Kirchman PA, Allen M & Jazwinski SM Suppressor analysis points to the subtle role of the LAG1 ceramide synthase gene in determining yeast longevity. Exp. Gerontol 39, 999–1009 (2004). - PubMed
-
- Jiang JC, Kirchman PA, Zagulski M, Hunt J & Jazwinski SM Homologs of the yeast longevity gene LAG1 in Caenorhabditis elegans and human. Genome Res. 8, 1259–1272 (1998). - PubMed
-
- Hannun YA & Obeid LM Principles of bioactive lipid signalling: lessons from sphingolipids. Nat. Rev. Mol. Cell Biol 9, 139–150 (2008). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

