PD-1 defines a distinct, functional, tissue-adapted state in Vδ1+ T cells with implications for cancer immunotherapy
- PMID: 38172341
- PMCID: PMC10965442
- DOI: 10.1038/s43018-023-00690-0
PD-1 defines a distinct, functional, tissue-adapted state in Vδ1+ T cells with implications for cancer immunotherapy
Abstract
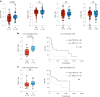
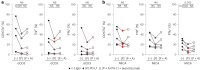
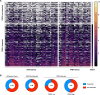
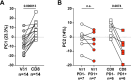
Checkpoint inhibition (CPI), particularly that targeting the inhibitory coreceptor programmed cell death protein 1 (PD-1), has transformed oncology. Although CPI can derepress cancer (neo)antigen-specific αβ T cells that ordinarily show PD-1-dependent exhaustion, it can also be efficacious against cancers evading αβ T cell recognition. In such settings, γδ T cells have been implicated, but the functional relevance of PD-1 expression by these cells is unclear. Here we demonstrate that intratumoral TRDV1 transcripts (encoding the TCRδ chain of Vδ1+ γδ T cells) predict anti-PD-1 CPI response in patients with melanoma, particularly those harboring below average neoantigens. Moreover, using a protocol yielding substantial numbers of tissue-derived Vδ1+ cells, we show that PD-1+Vδ1+ cells display a transcriptomic program similar to, but distinct from, the canonical exhaustion program of colocated PD-1+CD8+ αβ T cells. In particular, PD-1+Vδ1+ cells retained effector responses to TCR signaling that were inhibitable by PD-1 engagement and derepressed by CPI.
© 2024. The Author(s).
Conflict of interest statement
D.D., S.K. and M.L.I. were previously employed on a sponsored research agreement with Gamma Delta Therapeutics. D.D. and S.K. are currently employed on a sponsored research agreement with Takeda Pharmaceuticals (starting in February 2023). I.Z. was previously employed on a sponsored research agreement with Takeda Pharmaceuticals (February–July 2023). D.B. reports personal fees from NanoString and AstraZeneca and has a patent (PCT/GB2020/050221) issued on methods for cancer prognostication. A.H. receives laboratory research funding from Takeda Pharmaceuticals (starting in February 2023) and consults for eGenesis and Prokarium. Y.W. consults for PersonGen Biotherapeutics and E15 VC. The remaining authors declare no competing interests.
Figures








References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

