The eRF1 degrader SRI-41315 acts as a molecular glue at the ribosomal decoding center
- PMID: 38172604
- PMCID: PMC11253071
- DOI: 10.1038/s41589-023-01521-0
The eRF1 degrader SRI-41315 acts as a molecular glue at the ribosomal decoding center
Abstract
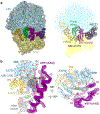
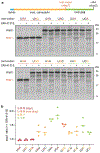
Translation termination is an essential cellular process, which is also of therapeutic interest for diseases that manifest from premature stop codons. In eukaryotes, translation termination requires eRF1, which recognizes stop codons, catalyzes the release of nascent proteins from ribosomes and facilitates ribosome recycling. The small molecule SRI-41315 triggers eRF1 degradation and enhances translational readthrough of premature stop codons. However, the mechanism of action of SRI-41315 on eRF1 and translation is not known. Here we report cryo-EM structures showing that SRI-41315 acts as a metal-dependent molecular glue between the N domain of eRF1 responsible for stop codon recognition and the ribosomal subunit interface near the decoding center. Retention of eRF1 on ribosomes by SRI-41315 leads to ribosome collisions, eRF1 ubiquitylation and a higher frequency of translation termination at near-cognate stop codons. Our findings reveal a new mechanism of release factor inhibition and additional implications for pharmacologically targeting eRF1.
© 2024. The Author(s), under exclusive licence to Springer Nature America, Inc.
Figures






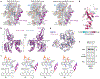



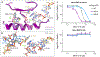
References
-
- Frolova L et al. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature 372, 701–703 (1994). - PubMed
Methods References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

