Methanolobus use unspecific methyltransferases to produce methane from dimethylsulphide in Baltic Sea sediments
- PMID: 38172958
- PMCID: PMC10762971
- DOI: 10.1186/s40168-023-01720-w
Methanolobus use unspecific methyltransferases to produce methane from dimethylsulphide in Baltic Sea sediments
Abstract
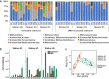
Background: In anoxic coastal and marine sediments, degradation of methylated compounds is the major route to the production of methane, a powerful greenhouse gas. Dimethylsulphide (DMS) is the most abundant biogenic organic sulphur compound in the environment and an abundant methylated compound leading to methane production in anoxic sediments. However, understanding of the microbial diversity driving DMS-dependent methanogenesis is limited, and the metabolic pathways underlying this process in the environment remain unexplored. To address this, we used anoxic incubations, amplicon sequencing, genome-centric metagenomics and metatranscriptomics of brackish sediments collected along the depth profile of the Baltic Sea with varying sulphate concentrations.
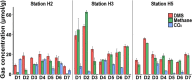
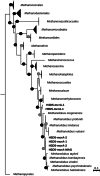
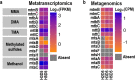
Results: We identified Methanolobus as the dominant methylotrophic methanogens in all our DMS-amended sediment incubations (61-99%) regardless of their sulphate concentrations. We also showed that the mtt and mta genes (trimethylamine- and methanol-methyltransferases) from Methanolobus were highly expressed when the sediment samples were incubated with DMS. Furthermore, we did not find mtsA and mtsB (methylsulphide-methyltransferases) in metatranscriptomes, metagenomes or in the Methanolobus MAGs, whilst mtsD and mtsF were found 2-3 orders of magnitude lower in selected samples.
Conclusions: Our study demonstrated that the Methanolobus genus is likely the key player in anaerobic DMS degradation in brackish Baltic Sea sediments. This is also the first study analysing the metabolic pathways of anaerobic DMS degradation in the environment and showing that methylotrophic methane production from DMS may not require a substrate-specific methyltransferase as was previously accepted. This highlights the versatility of the key enzymes in methane production in anoxic sediments, which would have significant implications for the global greenhouse gas budget and the methane cycle. Video Abstract.
Keywords: Dimethylsulphide; Metagenomics; Metatranscriptomics; Methanogenesis.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Watts SF. The mass budgets of carbonyl sulfide, dimethyl sulfide, carbon disulfide and hydrogen sulfide. Atmos Environ. 2000;34(5):761–79. doi: 10.1016/S1352-2310(99)00342-8. - DOI
-
- Charlson RJ, Lovelock JE, Andreae MO, Warren SG. Oceanic phytoplankton, atmospheric sulphur, cloud albedo and climate. Nature. 1987;326:655–661. doi: 10.1038/326655a0. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

