Interpretable machine learning model for early prediction of 28-day mortality in ICU patients with sepsis-induced coagulopathy: development and validation
- PMID: 38172962
- PMCID: PMC10763177
- DOI: 10.1186/s40001-023-01593-7
Interpretable machine learning model for early prediction of 28-day mortality in ICU patients with sepsis-induced coagulopathy: development and validation
Abstract
Objective: Sepsis-induced coagulopathy (SIC) is extremely common in individuals with sepsis, significantly associated with poor outcomes. This study attempted to develop an interpretable and generalizable machine learning (ML) model for early predicting the risk of 28-day death in patients with SIC.
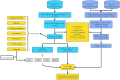
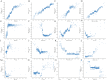
Methods: In this retrospective cohort study, we extracted SIC patients from the Medical Information Mart for Intensive Care III (MIMIC-III), MIMIC-IV, and eICU-CRD database according to Toshiaki Iba's scale. And the overlapping in the MIMIC-IV was excluded for this study. Afterward, only the MIMIC-III cohort was randomly divided into the training set, and the internal validation set according to the ratio of 7:3, while the MIMIC-IV and eICU-CRD databases were considered the external validation sets. The predictive factors for 28-day mortality of SIC patients were determined using recursive feature elimination combined with tenfold cross-validation (RFECV). Then, we constructed models using ML algorithms. Multiple metrics were used for evaluation of performance of the models, including the area under the receiver operating characteristic curve (AUROC), area under the precision recall curve (AUPRC), accuracy, sensitivity, specificity, negative predictive value, positive predictive value, recall, and F1 score. Finally, Shapley Additive Explanations (SHAP), Local Interpretable Model-Agnostic Explanations (LIME) were employed to provide a reasonable interpretation for the prediction results.
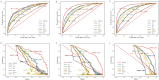
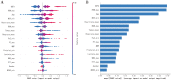
Results: A total of 3280, 2798, and 1668 SIC patients were screened from MIMIC-III, MIMIC-IV, and eICU-CRD databases, respectively. Seventeen features were selected to construct ML prediction models. XGBoost had the best performance in predicting the 28-day mortality of SIC patients, with AUC of 0.828, 0.913 and 0.923, the AUPRC of 0.807, 0.796 and 0.921, the accuracy of 0.785, 0.885 and 0.891, the F1 scores were 0.63, 0.69 and 0.70 in MIMIC-III (internal validation set), MIMIC-IV, and eICU-CRD databases. The importance ranking and SHAP analyses showed that initial SOFA score, red blood cell distribution width (RDW), and age were the top three critical features in the XGBoost model.
Conclusions: We developed an optimal and explainable ML model to predict the risk of 28-day death of SIC patients 28-day death risk. Compared with conventional scoring systems, the XGBoost model performed better. The model established will have the potential to improve the level of clinical practice for SIC patients.
Keywords: Gradient boosting decision tree; Local interpretable model-agnostic explanations; Machine learning; Sepsis induced coagulopathy; Shapley additive explanations.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
Interpretable machine learning for 28-day all-cause in-hospital mortality prediction in critically ill patients with heart failure combined with hypertension: A retrospective cohort study based on medical information mart for intensive care database-IV and eICU databases.Front Cardiovasc Med. 2022 Oct 12;9:994359. doi: 10.3389/fcvm.2022.994359. eCollection 2022. Front Cardiovasc Med. 2022. PMID: 36312291 Free PMC article.
-
A Machine-Learning Approach for Dynamic Prediction of Sepsis-Induced Coagulopathy in Critically Ill Patients With Sepsis.Front Med (Lausanne). 2021 Jan 21;7:637434. doi: 10.3389/fmed.2020.637434. eCollection 2020. Front Med (Lausanne). 2021. PMID: 33553224 Free PMC article.
-
A machine learning model for robust prediction of sepsis-induced coagulopathy in critically ill patients with sepsis.Front Cell Infect Microbiol. 2025 Jun 6;15:1579558. doi: 10.3389/fcimb.2025.1579558. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40546281 Free PMC article.
-
Interpretable machine learning model to predict surgical difficulty in laparoscopic resection for rectal cancer.Front Oncol. 2024 Feb 6;14:1337219. doi: 10.3389/fonc.2024.1337219. eCollection 2024. Front Oncol. 2024. PMID: 38380369 Free PMC article. Review.
-
Machine Learning Models in Sepsis Outcome Prediction for ICU Patients: Integrating Routine Laboratory Tests-A Systematic Review.Biomedicines. 2024 Dec 19;12(12):2892. doi: 10.3390/biomedicines12122892. Biomedicines. 2024. PMID: 39767798 Free PMC article. Review.
Cited by
-
An interpretable machine learning model for predicting in-hospital mortality in ICU patients with ventilator-associated pneumonia.PLoS One. 2025 Jan 7;20(1):e0316526. doi: 10.1371/journal.pone.0316526. eCollection 2025. PLoS One. 2025. PMID: 39774553 Free PMC article.
-
Early prognosis prediction for non-variceal upper gastrointestinal bleeding in the intensive care unit: based on interpretable machine learning.Eur J Med Res. 2024 Aug 31;29(1):442. doi: 10.1186/s40001-024-02005-0. Eur J Med Res. 2024. PMID: 39217369 Free PMC article.
-
A multicenter study on developing a prognostic model for severe fever with thrombocytopenia syndrome using machine learning.Front Microbiol. 2025 Mar 19;16:1557922. doi: 10.3389/fmicb.2025.1557922. eCollection 2025. Front Microbiol. 2025. PMID: 40177493 Free PMC article.
-
Interpretable machine learning model for early prediction of delirium in elderly patients following intensive care unit admission: a derivation and validation study.Front Med (Lausanne). 2024 May 17;11:1399848. doi: 10.3389/fmed.2024.1399848. eCollection 2024. Front Med (Lausanne). 2024. PMID: 38828233 Free PMC article.
-
Predictive value of peripheral blood indicators plus procalcitonin clearance rate for mortality in cancer patients with sepsis.Am J Cancer Res. 2024 Dec 15;14(12):5839-5850. doi: 10.62347/NKOL2327. eCollection 2024. Am J Cancer Res. 2024. PMID: 39803656 Free PMC article.
References
-
- Rudd KE, Johnson SC, Agesa KM, et al. Global, regional, and national sepsis incidence and mortality, 1990–2017: Editorials Copyright © 2021 by the Society of Critical Care Medicine and Wolters Kluwer Health, Inc. All Rights Reserved.Critical Care Medicine www.ccmjournal.org 863 analysis for the global burden of disease study. Lancet. 2020;395:200–211. doi: 10.1016/S0140-6736(19)32989-7. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

