Transcriptional profiling of human cartilage endplate cells identifies novel genes and cell clusters underlying degenerated and non-degenerated phenotypes
- PMID: 38173036
- PMCID: PMC10763221
- DOI: 10.1186/s13075-023-03220-6
Transcriptional profiling of human cartilage endplate cells identifies novel genes and cell clusters underlying degenerated and non-degenerated phenotypes
Abstract
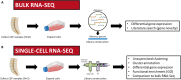
Background: Low back pain is a leading cause of disability worldwide and is frequently attributed to intervertebral disc (IVD) degeneration. Though the contributions of the adjacent cartilage endplates (CEP) to IVD degeneration are well documented, the phenotype and functions of the resident CEP cells are critically understudied. To better characterize CEP cell phenotype and possible mechanisms of CEP degeneration, bulk and single-cell RNA sequencing of non-degenerated and degenerated CEP cells were performed.
Methods: Human lumbar CEP cells from degenerated (Thompson grade ≥ 4) and non-degenerated (Thompson grade ≤ 2) discs were expanded for bulk (N=4 non-degenerated, N=4 degenerated) and single-cell (N=1 non-degenerated, N=1 degenerated) RNA sequencing. Genes identified from bulk RNA sequencing were categorized by function and their expression in non-degenerated and degenerated CEP cells were compared. A PubMed literature review was also performed to determine which genes were previously identified and studied in the CEP, IVD, and other cartilaginous tissues. For single-cell RNA sequencing, different cell clusters were resolved using unsupervised clustering and functional annotation. Differential gene expression analysis and Gene Ontology, respectively, were used to compare gene expression and functional enrichment between cell clusters, as well as between non-degenerated and degenerated CEP samples.
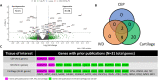
Results: Bulk RNA sequencing revealed 38 genes were significantly upregulated and 15 genes were significantly downregulated in degenerated CEP cells relative to non-degenerated cells (|fold change| ≥ 1.5). Of these, only 2 genes were previously studied in CEP cells, and 31 were previously studied in the IVD and other cartilaginous tissues. Single-cell RNA sequencing revealed 11 unique cell clusters, including multiple chondrocyte and progenitor subpopulations with distinct gene expression and functional profiles. Analysis of genes in the bulk RNA sequencing dataset showed that progenitor cell clusters from both samples were enriched in "non-degenerated" genes but not "degenerated" genes. For both bulk- and single-cell analyses, gene expression and pathway enrichment analyses highlighted several pathways that may regulate CEP degeneration, including transcriptional regulation, translational regulation, intracellular transport, and mitochondrial dysfunction.
Conclusions: This thorough analysis using RNA sequencing methods highlighted numerous differences between non-degenerated and degenerated CEP cells, the phenotypic heterogeneity of CEP cells, and several pathways of interest that may be relevant in CEP degeneration.
Keywords: Bulk RNA-Seq; Cartilage endplate; Degeneration; Human; Intervertebral disc; Single-cell RNA-Seq.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







References
-
- James SL, et al. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2018;392:1789–1858. doi: 10.1016/S0140-6736(18)32279-7. - DOI - PMC - PubMed
-
- Andersson G. Epidemiological features of chronic low-back pain. 1999. - PubMed
-
- Katz JN. Lumbar disc disorders and low-back pain: socioeconomic factors and consequences. J Bone Joint Surg Am. 2006;88:21–24. - PubMed
-
- Ferreira ML, et al. Global, regional, and national burden of low back pain, 1990–2020, its attributable risk factors, and projections to 2050: a systematic analysis of the Global Burden of Disease Study 2021. Lancet Rheumatol. 2023;5:e316–e329. doi: 10.1016/S2665-9913(23)00098-X. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

